8APM
 
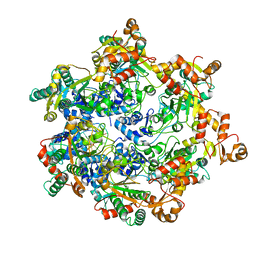 | | Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C1 | | Descriptor: | DNA (5'-D(P*CP*CP*GP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*TP*TP*CP*GP*G)-3'), Primase D5 | | Authors: | Burmeister, W.P, Hutin, S, Ling, W.L, Grimm, C, Schoehn, G. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction.
Viruses, 14, 2022
|
|
6RIE
 
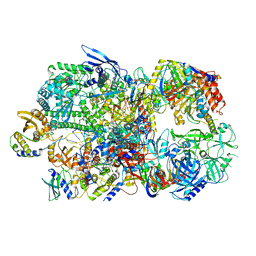 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase co-transcriptional capping complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6RID
 
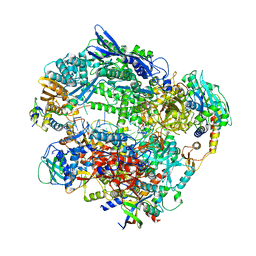 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase elongation complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
2VY2
 
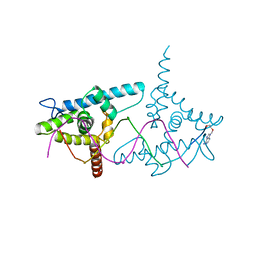 | | Structure of LEAFY transcription factor from Arabidopsis thaliana in complex with DNA from AG-I promoter | | Descriptor: | 5'-D(*AP*TP*TP*TP*AP*AP*TP*CP*CP*AP *AP*TP*GP*GP*TP*TP*AP*CP*AP*A)-3', PROTEIN LEAFY | | Authors: | Hames, C, Ptchelkine, D, Grimm, C, Thevenon, E, Moyroud, E, Gerard, F, Martiel, J.L, Benlloch, R, Parcy, F, Muller, C.W. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Leafy Floral Switch Function and Similarity with Helix-Turn-Helix Proteins.
Embo J., 27, 2008
|
|
2VY1
 
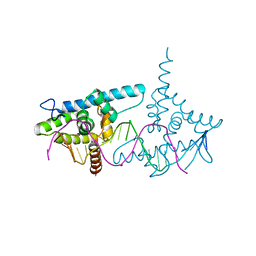 | | Structure of LEAFY transcription factor from Arabidopsis thaliana in complex with DNA from AP1 promoter | | Descriptor: | 5'-D(*TP*TP*AP*CP*GP*GP*AP*CP*CP*AP *CP*TP*GP*GP*TP*CP*CP*TP*TP*CP)-3', PROTEIN LEAFY | | Authors: | Hames, C, Ptchelkine, D, Grimm, C, Thevenon, E, Moyroud, E, Gerard, F, Martiel, J.L, Benlloch, R, Parcy, F, Muller, C.W. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Basis for Leafy Floral Switch Function and Similarity with Helix-Turn-Helix Proteins.
Embo J., 27, 2008
|
|
7O6R
 
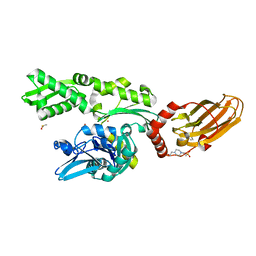 | |
7ODI
 
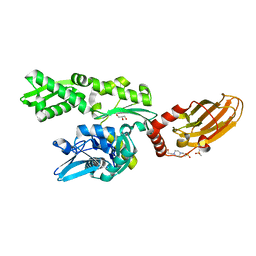 | |
7ODB
 
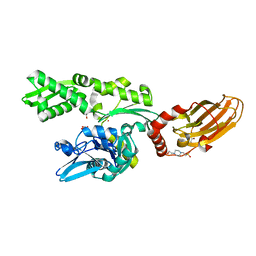 | | Crystal structure of bovine Hsc70(aa1-554)E213A/D214A in complex with triazine-derivative | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-methyl-5-sulfanylidene-2H-1,2,4-triazin-3-one, GLYCEROL, ... | | Authors: | Zehe, M, Grimm, C, Sotriffer, C. | | Deposit date: | 2021-04-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Combined In-Solution Fragment Screening and Crystallographic Binding-Mode Analysis with a Two-Domain Hsp70 Construct.
Acs Chem.Biol., 2024
|
|
7ODD
 
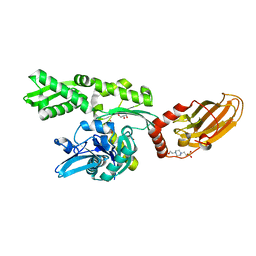 | |
7BB3
 
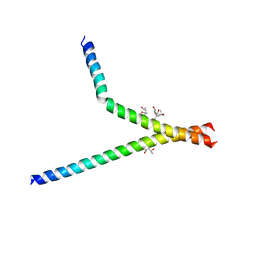 | | Structure of S. pombe YG-box oligomer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Survival motor neuron-like protein 1,Survival motor neuron-like protein 1 | | Authors: | Veepaschit, J, Grimm, C, Fischer, U. | | Deposit date: | 2020-12-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Identification and structural analysis of the Schizosaccharomyces pombe SMN complex.
Nucleic Acids Res., 49, 2021
|
|
7BBL
 
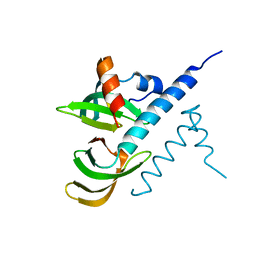 | |
8C8H
 
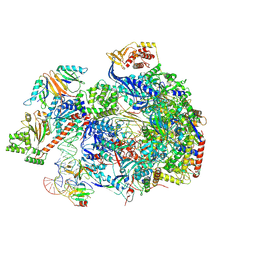 | |
8QAM
 
 | |
8OUO
 
 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 32, 2024
|
|
2X6U
 
 | | Crystal structure of human TBX5 in the DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX5 | | Authors: | Stirnimann, C.U, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
