4QIQ
 
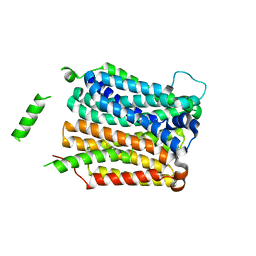 | | Crystal structure of D-xylose-proton symporter | | Descriptor: | D-xylose-proton symporter, ZINC ION | | Authors: | Wisedchaisri, G, Park, M, Iadanza, M.G, Zheng, H, Gonen, T. | | Deposit date: | 2014-06-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE.
Nat Commun, 5, 2014
|
|
6M7M
 
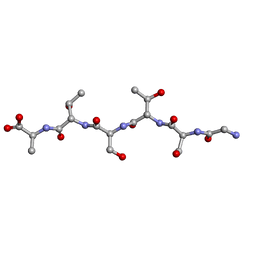 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6M9J
 
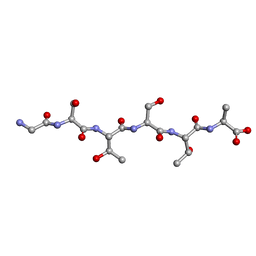 | | Racemic-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6N4U
 
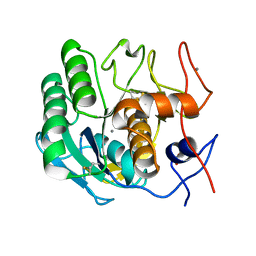 | | MicroED structure of Proteinase K at 2.75A resolution from a single milled crystal. | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2018-11-20 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.75 Å) | | Cite: | Collection of Continuous Rotation MicroED Data from Ion Beam-Milled Crystals of Any Size.
Structure, 27, 2019
|
|
6NB9
 
 | | Amyloid-Beta (20-34) with L-isoaspartate 23 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Boyer, D.R, Zee, C.T, Richards, L.S, Cascio, D, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-08-07 | | Last modified: | 2022-09-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
6OIZ
 
 | | Amyloid-Beta (20-34) wild type | | Descriptor: | Amyloid beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Zee, C.T, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
3J07
 
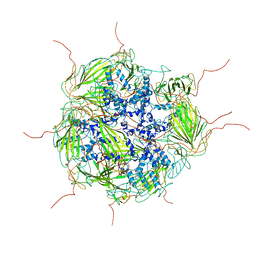 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3J7B
 
 | | Catalase solved at 3.2 Angstrom resolution by MicroED | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nannenga, B.L, Shi, D, Hattne, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Structure of catalase determined by MicroED.
Elife, 3, 2014
|
|
3J41
 
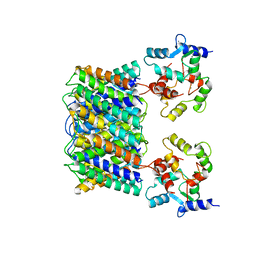 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3J4G
 
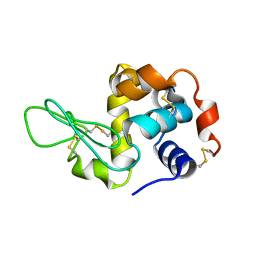 | |
3J4Q
 
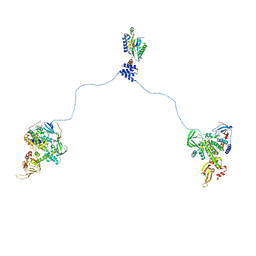 | |
3J4R
 
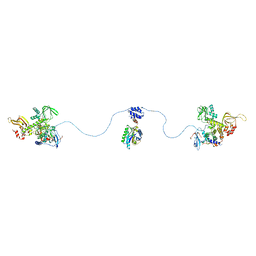 | |
3J6K
 
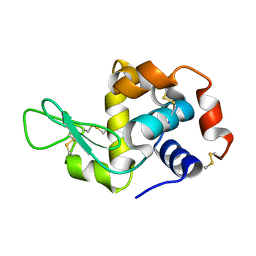 | |
8W12
 
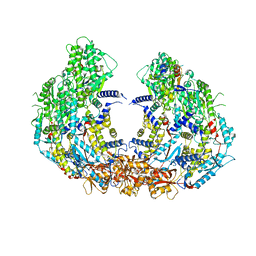 | | Cryo-EM structure of VP3-VP6 heterohexamer | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1C
 
 | | Cryo-EM structure of BTV pre-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1S
 
 | | Cryo-EM structure of BTV pre-core | | Descriptor: | Core protein VP3, RNA-directed RNA polymerase | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1I
 
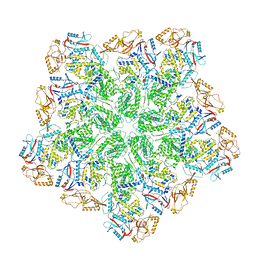 | | Cryo-EM structure of BTV subcore | | Descriptor: | Core protein VP3 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1R
 
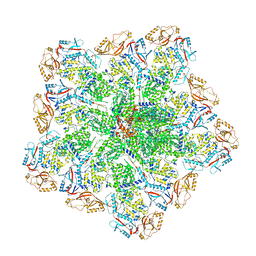 | | Cryo-EM structure of BTV core | | Descriptor: | Core protein VP3, RNA-directed RNA polymerase | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1O
 
 | | Cryo-EM structure of BTV virion | | Descriptor: | Core protein VP3, Outer capsid protein VP2, RNA-1, ... | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W19
 
 | | Cryo-EM structure of BTV star-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
7SKX
 
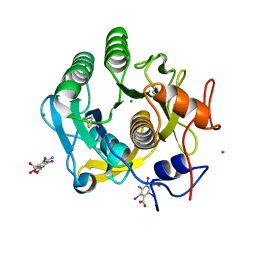 | | Ab initio structure of proteinase K from electron-counted MicroED data | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Hattne, J, Gonen, T. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Ab initio phasing macromolecular structures using electron-counted MicroED data.
Nat.Methods, 19, 2022
|
|
7SKW
 
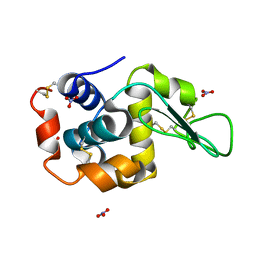 | | Ab initio structure of triclinic lysozyme from electron-counted MicroED data | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Hattne, J, Gonen, T. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.87 Å) | | Cite: | Ab initio phasing macromolecular structures using electron-counted MicroED data.
Nat.Methods, 19, 2022
|
|
7SW4
 
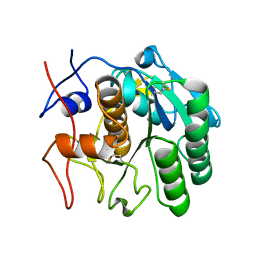 | | MicroED structure of proteinase K from a 540 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW9
 
 | | MicroED structure of proteinase K from a 170 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW2
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
