4G3O
 
 | |
7SOT
 
 | |
7SOO
 
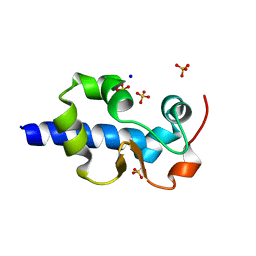 | | LaM domain of human LARP1 | | Descriptor: | Isoform 2 of La-related protein 1, SODIUM ION, SULFATE ION | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOQ
 
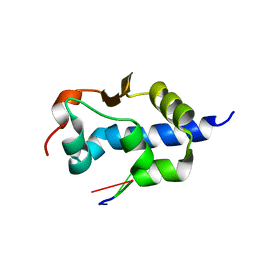 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | Isoform 2 of La-related protein 1, RNA (5'-R(*AP*AP*A)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOV
 
 | |
7SOR
 
 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoform 2 of La-related protein 1, ... | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOP
 
 | |
7SOU
 
 | |
7SOS
 
 | | LaM domain of human LARP1 in complex with AAAA RNA | | Descriptor: | Isoform 2 of La-related protein 1, POTASSIUM ION, RNA (5'-R(*AP*AP*AP*A)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOW
 
 | | LaM domain of human LARP1 in complex with UUUUUU | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of La-related protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of 3'-end RNA recognition by LARP1
To Be Published
|
|
4IOT
 
 | | High-resolution Structure of Triosephosphate isomerase from E. coli | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Vinaik, R, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-01-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triosephosphate isomerase is a common crystallization contaminant of soluble His-tagged proteins produced in Escherichia coli.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JU5
 
 | |
7US1
 
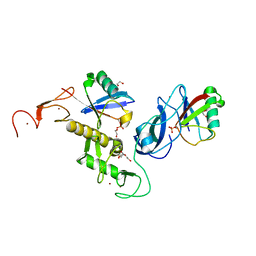 | | Structure of parkin (R0RB) bound to two phospho-ubiquitin molecules | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Fakih, R, Sauve, V, Gehring, K. | | Deposit date: | 2022-04-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structure of the second phosphoubiquitin-binding site in parkin.
J.Biol.Chem., 298, 2022
|
|
4I6X
 
 | |
4JRD
 
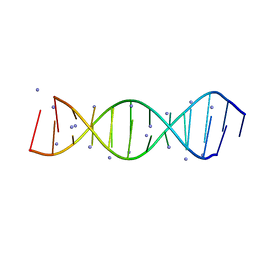 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2OOB
 
 | |
5TDB
 
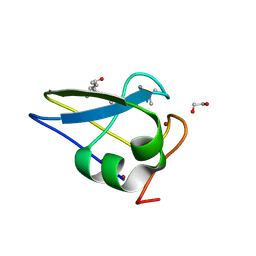 | | Crystal structure of the human UBR-box domain from UBR2 in complex with asymmetrically double methylated arginine peptide | | Descriptor: | 1,2-ETHANEDIOL, DA2-ILE-PHE-SER peptide, E3 ubiquitin-protein ligase UBR2, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
2QHO
 
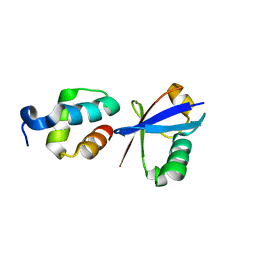 | |
5TDC
 
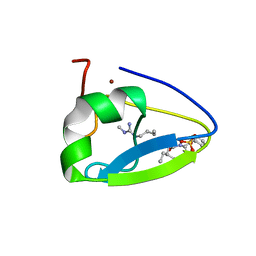 | | Crystal structure of the human UBR-box domain from UBR1 in complex with monomethylated arginine peptide. | | Descriptor: | E3 ubiquitin-protein ligase UBR1, NMM-ILE-PHE-SER peptide, SULFATE ION, ... | | Authors: | Kozlov, G, Munoz-Escobar, J, Matta-Camacho, E, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
2OOA
 
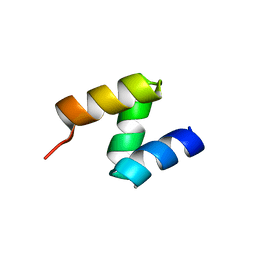 | |
5TGG
 
 | |
5TDD
 
 | | Human UBR-box from UBR2 in complex with HIFS peptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UBR2, HIS-ILE-PHE-SER peptide, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TSR
 
 | |
2OO9
 
 | |
5BTW
 
 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
