1QNW
 
 | | lectin II from Ulex europaeus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-10-22 | | Release date: | 1999-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1Q8Q
 
 | | Pterocarpus angolensis lectin (PAL) in complex with the dimannoside Man(alpha1-4)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-4)-methyl alpha-D-mannopyranoside, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
1QOO
 
 | | lectin UEA-II complexed with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1Q8V
 
 | | Pterocarpus angolensis lectin (PAL) in complex with the trimannoside [Man(Alpha1-3)]Man(alpha1-6)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
4BWO
 
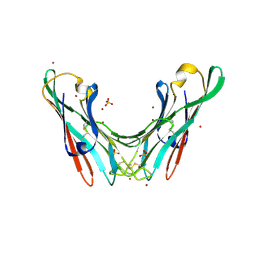 | | The FedF adhesin from entrrotoxigenic Escherichia coli is a sulfate- binding lectin | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Lonardi, E, Moonens, K, Buts, L, de Boer, A.R, Olsson, J.D.M, Weiss, M.S, Fabre, E, Guerardel, Y, Deelder, A.M, Oscarson, S, Wuhrer, M, Bouckaert, J. | | Deposit date: | 2013-07-03 | | Release date: | 2013-08-28 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia Coli
Biology, 2, 2013
|
|
7QUO
 
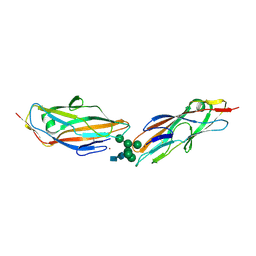 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
2BS8
 
 | | Crystal structure of F17b-G in complex with N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESIN | | Authors: | Buts, L, Wellens, A, VanMolle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, DeGreve, H, Bouckaert, J. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6HHU
 
 | |
6QX4
 
 | |
4ZM0
 
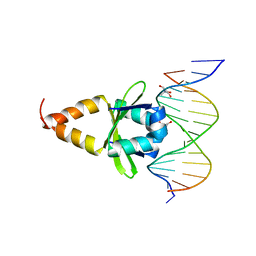 | |
4ZM2
 
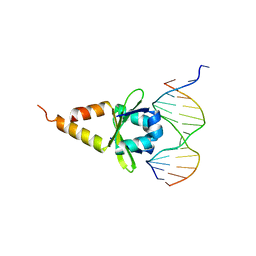 | |
4ZLX
 
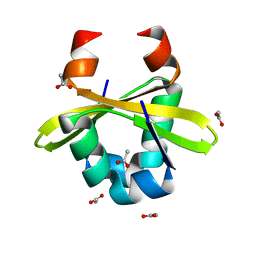 | |
5AFO
 
 | |
2MF2
 
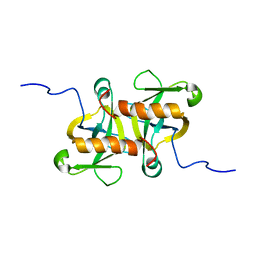 | |
7BHD
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
3G7Z
 
 | | CcdB dimer in complex with two C-terminal CcdA domains | | Descriptor: | Cytotoxic protein ccdB, Protein ccdA | | Authors: | De Jonge, N, Loris, R, Garcia-Pino, A, Buts, L. | | Deposit date: | 2009-02-11 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
Mol.Cell, 35, 2009
|
|
3HPW
 
 | | CcdB dimer in complex with one C-terminal CcdA domain | | Descriptor: | Cytotoxic protein ccdB, DI(HYDROXYETHYL)ETHER, Protein ccdA, ... | | Authors: | De Jonge, N, Loris, R, Garcia-Pino, A, Buts, L. | | Deposit date: | 2009-06-05 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
Mol.Cell, 35, 2009
|
|
3DD9
 
 | | Structure of DocH66Y dimer | | Descriptor: | Death on curing protein | | Authors: | Garcia-Pino, A, Loris, R. | | Deposit date: | 2008-06-05 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The intrinsically disordered domain of the antitoxin Phd chaperones the toxin Doc against irreversible inactivation and misfolding
J. Biol. Chem., 289, 2014
|
|
5CKF
 
 | | E. coli MazF E24A form I | | Descriptor: | Endoribonuclease MazF, SODIUM ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKB
 
 | | E. coli MazF form II | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKE
 
 | | E.coli MazF E24A form IIa | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Endoribonuclease MazF, SULFATE ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKD
 
 | | E. coli MazF E24A form III | | Descriptor: | Endoribonuclease MazF, SODIUM ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CO7
 
 | | E. coli MazF form III | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CK9
 
 | | E. coli MazF form I | | Descriptor: | Endoribonuclease MazF, PENTAETHYLENE GLYCOL | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKH
 
 | | E. coli MazF E24A form IIb | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
