3DXJ
 
 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
1QE1
 
 | | CRYSTAL STRUCTURE OF 3TC-RESISTANT M184I MUTANT OF HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | REVERSE TRANSCRIPTASE, SUBUNIT P51, SUBUNIT P66 | | Authors: | Sarafianos, S.G, Das, K, Ding, J, Hughes, S.H, Arnold, E. | | Deposit date: | 1999-07-12 | | Release date: | 1999-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4W9S
 
 | | 2-(4-(1H-tetrazol-5-yl)phenyl)-5-hydroxypyrimidin-4(3H)-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 5-hydroxy-2-[4-(1H-tetrazol-5-yl)phenyl]pyrimidin-4(3H)-one, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phenyl Substituted 4-Hydroxypyridazin-3(2H)-ones and 5-Hydroxypyrimidin-4(3H)-ones: Inhibitors of Influenza A Endonuclease.
J.Med.Chem., 57, 2014
|
|
7LQU
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | Deposit date: | 2021-02-15 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
3KLF
 
 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLE
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4M5V
 
 | | Influenza 2009 H1N1 endonuclease with 100 millimolar calcium | | Descriptor: | CALCIUM ION, Polymerase PA, SULFATE ION | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
4MK1
 
 | | 5-bromopyridine-2,3-diol bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-3-hydroxypyridin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
4MK5
 
 | | 6-(3-methoxyphenyl)pyridine-2,3-diol bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-(3-methoxyphenyl)pyridin-2(5H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
4MK2
 
 | | 3-(5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzonitrile bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzonitrile, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
7Z2E
 
 | |
7Z2H
 
 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2G
 
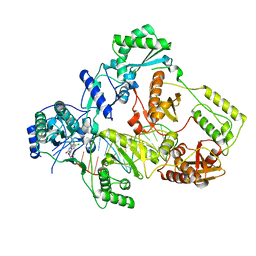 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z24
 
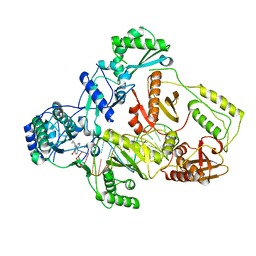 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-mer), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z29
 
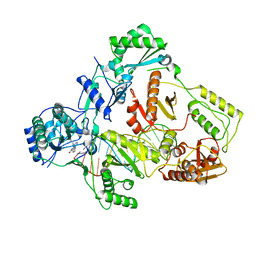 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2D
 
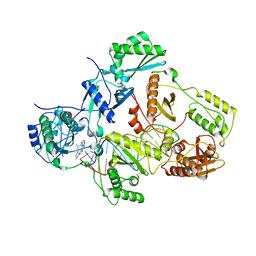 | |
8Q63
 
 | | Cryo-EM structure of IC8', a second state of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6ANQ
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.5 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-14 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
6DV9
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 4nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVC
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 6nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVD
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) with 6 nt spacer and bromine labelled in position "-11 | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.899 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVB
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 5nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVE
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF selenomethionine-labelled sigma factor L) with 6 nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*GP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
7OZ2
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA showing a transient P-pocket | | Descriptor: | CADMIUM ION, DNA (28-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
