6MJ8
 
 | |
6P8O
 
 | | Structure of P. aeruginosa ATCC27853 HORMA2-deltaC | | Descriptor: | CHLORIDE ION, HORMA domain containing protein, NICKEL (II) ION | | Authors: | Ye, Q, Corbett, K.D, Lau, R.K. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6MJB
 
 | |
8TYX
 
 | |
8TYY
 
 | |
8TZ0
 
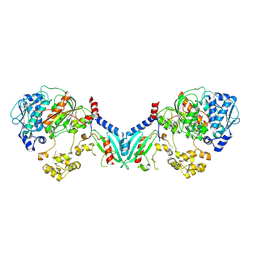 | | Structure of a bacterial E1-E2-Ubl complex (form 1) | | Descriptor: | E1(BilD), E2(BilB), Ubl(BilA), ... | | Authors: | Ye, Q, Chambers, L.R, Corbett, K.D. | | Deposit date: | 2023-08-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A eukaryotic-like ubiquitination system in bacterial antiviral defence.
Nature, 631, 2024
|
|
8TYZ
 
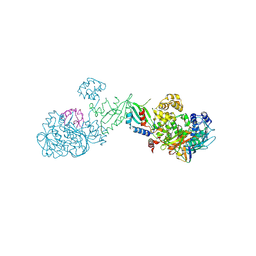 | | Structure of a bacterial E1-E2-Ubl complex (form 2) | | Descriptor: | E1(BilD), E2(BilB), Ubl(BilA), ... | | Authors: | Ye, Q, Chambers, L.R, Corbett, K.D. | | Deposit date: | 2023-08-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A eukaryotic-like ubiquitination system in bacterial antiviral defence.
Nature, 631, 2024
|
|
9C5G
 
 | |
8V48
 
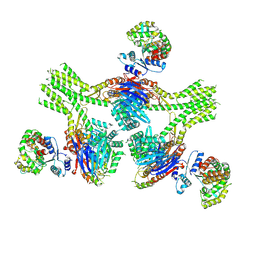 | | CryoEM structure of AriA-AriB complex (Form III) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
8V47
 
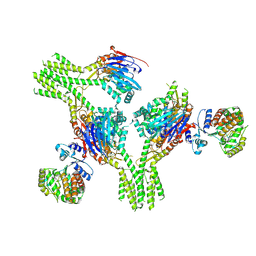 | | CryoEM structure of AriA-AriB complex (Form II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
8V46
 
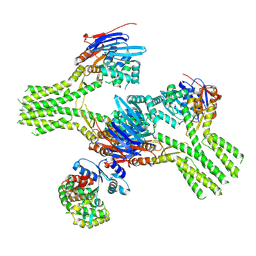 | | CryoEM structure of AriA-AriB complex (Form I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB, ... | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
8V49
 
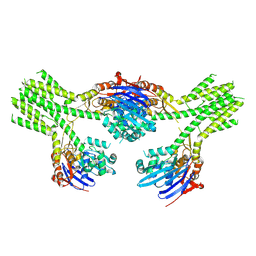 | |
8V45
 
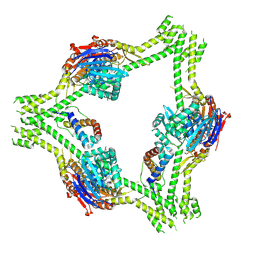 | | CryoEM structure of AriA-Ocr complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, Protein Ocr | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
2G2U
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
6UXF
 
 | | Structure of V. metoecus NucC, hexamer form | | Descriptor: | Vibrio meotecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6UXG
 
 | | Structure of V. metoecus NucC, trimer form | | Descriptor: | SULFATE ION, Vibrio metoecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
4TRK
 
 | | Structure of C. elegans HIM-3 | | Descriptor: | C. elegans HIM-3 | | Authors: | Rosenberg, S.C, Corbett, K.D. | | Deposit date: | 2014-06-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins.
Dev.Cell, 31, 2014
|
|
4TZL
 
 | |
4TZS
 
 | |
4TZQ
 
 | |
4TZO
 
 | |
4TZJ
 
 | |
4TZM
 
 | |
4TZN
 
 | | Structure of HTP-2 bound to HTP-3 motif-6 | | Descriptor: | Protein HTP-2, Protein HTP-3 | | Authors: | Rosenberg, S.C, Corbett, K.D. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins.
Dev.Cell, 31, 2014
|
|
2G2W
 
 | | Crystal Structure of the SHV D104K Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
