6Z3R
 
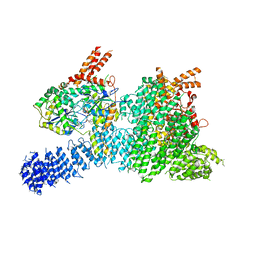 | | Structure of SMG1-8-9 kinase complex bound to UPF1-LSQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Langer, L.M, Gat, Y, Conti, E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of substrate-bound SMG1-8-9 kinase complex reveals molecular basis for phosphorylation specificity.
Elife, 9, 2020
|
|
4XR7
 
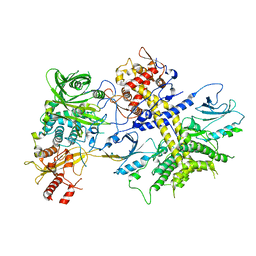 | | Structure of the Saccharomyces cerevisiae PAN2-PAN3 core complex | | Descriptor: | PAB-dependent poly(A)-specific ribonuclease subunit PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN3 | | Authors: | Schafer, I.B, Rode, M, Bonneau, F, Schussler, S, Conti, E. | | Deposit date: | 2015-01-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Nat. Struct. Mol. Biol., 21, 2014
|
|
7AQO
 
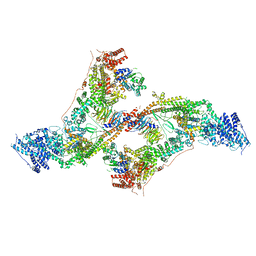 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APX
 
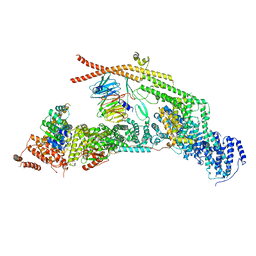 | | yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2,Tho2, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
1FT8
 
 | |
4WFD
 
 | | Structure of the Rrp6-Rrp47-Mtr4 interaction | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex exonuclease RRP6, Exosome complex protein LRP1, ... | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
1FO1
 
 | |
4Q8H
 
 | |
5LXR
 
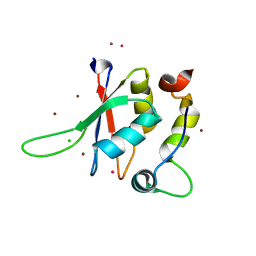 | | Structure of the minimal RBM7 - ZCCHC8 Complex | | Descriptor: | BROMIDE ION, CHLORIDE ION, RNA-binding protein 7, ... | | Authors: | Falk, S, Finogenova, K, Benda, C, Conti, E. | | Deposit date: | 2016-09-22 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors.
Nat Commun, 7, 2016
|
|
5LXY
 
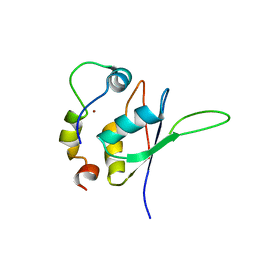 | | Structure of the minimal RBM7 - ZCCHC8 Complex | | Descriptor: | BROMIDE ION, RNA-binding protein 7, Zinc finger CCHC domain-containing protein 8 | | Authors: | Falk, S, Finogenova, K, Benda, C, Conti, E. | | Deposit date: | 2016-09-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors.
Nat Commun, 7, 2016
|
|
4Q8G
 
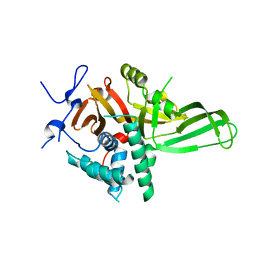 | |
6H25
 
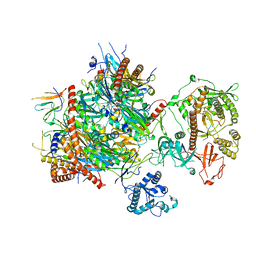 | | Human nuclear RNA exosome EXO-10-MPP6 complex | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Gerlach, P, Schuller, J.M, Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2018-07-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Distinct and evolutionary conserved structural features of the human nuclear exosome complex.
Elife, 7, 2018
|
|
2BR2
 
 | | RNase PH core of the archaeal exosome | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX EXONUCLEASE 1, EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | lorentzen, E, Fribourg, S, Conti, E. | | Deposit date: | 2005-04-30 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Archaeal Exosome Core is a Hexameric Ring Structure with Three Catalytic Subunits.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4TXD
 
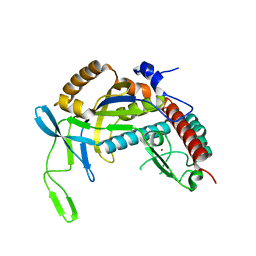 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
6R5K
 
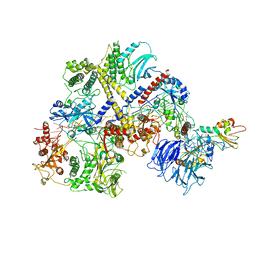 | |
6RO1
 
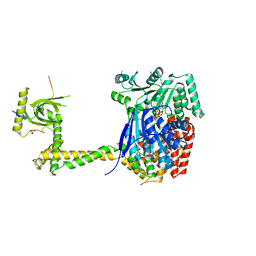 | | X-ray crystal structure of the MTR4 NVL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Lingaraju, M, Langer, L.M, Basquin, J, Falk, S, Conti, E. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs.
Nat Commun, 10, 2019
|
|
9G8N
 
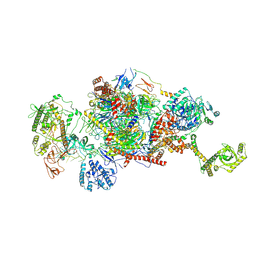 | | 80S-bound human Ski2-exosome complex | | Descriptor: | CrPV-IRES RNA, DIS3-like exonuclease 1, Exosome complex component CSL4, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8Q
 
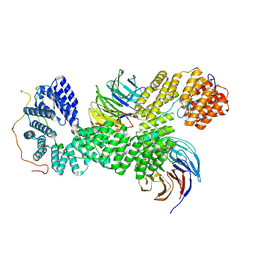 | | 40S-bound human SKI238 complex in the open state (Gatekeeping module) | | Descriptor: | Helicase SKI2W, Superkiller complex protein 3, WD repeat-containing protein 61 | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8P
 
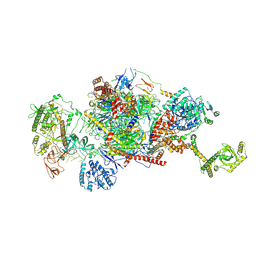 | | 40S-bound human SKI2-exosome complex | | Descriptor: | CrPV-IRES RNA, DIS3-like exonuclease 1, Exosome complex component CSL4, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8R
 
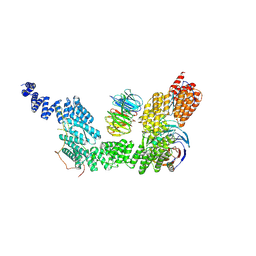 | | human SKI7-SKI238 complex in the open state | | Descriptor: | Isoform 2 of HBS1-like protein, Superkiller complex protein 2, Superkiller complex protein 3, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
2C39
 
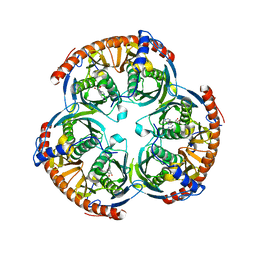 | | RNase PH core of the archaeal exosome in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
9G8O
 
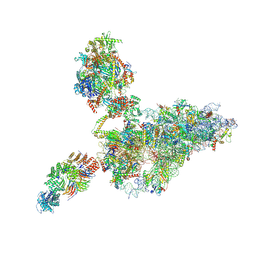 | | human 40S ribosome bound by a SKI238-exosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8M
 
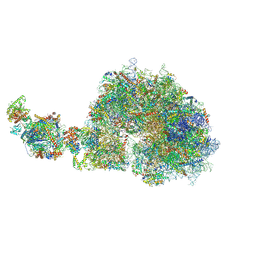 | | human 80S ribosome bound by a SKI2-exosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
1OL7
 
 | |
