5D5R
 
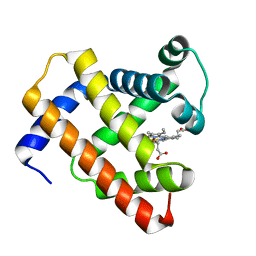 | | Horse-heart myoglobin - deoxy state | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Barends, T, Schlichting, I. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6PGK
 
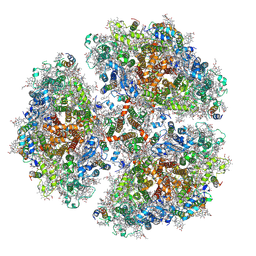 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Gisriel, C, Fromme, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
8FZW
 
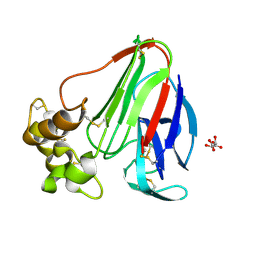 | | Thaumatin crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-01-30 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
8SCY
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7CWC
 
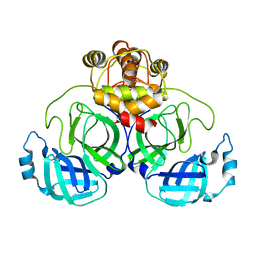 | |
7CWB
 
 | |
5G0Z
 
 | | Structure of native granulovirus polyhedrin determined using an X-ray free-electron laser | | Descriptor: | GRANULIN | | Authors: | Gati, C, Bunker, R.D, Oberthur, D, Metcalf, P, Henry, C. | | Deposit date: | 2016-03-23 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G3X
 
 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
9GXB
 
 | |
9GXC
 
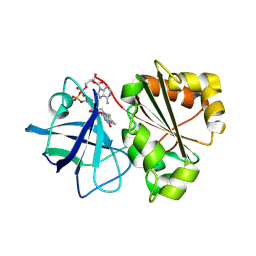 | |
4RW2
 
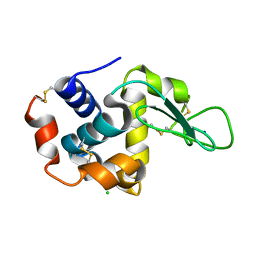 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: refocused beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Barends, T, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
7VK0
 
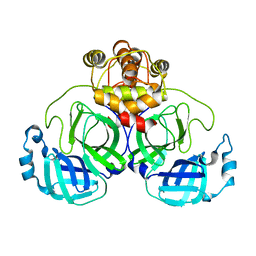 | |
7VK1
 
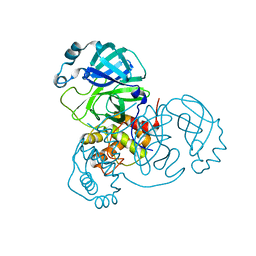 | | Crystal Structure of SARS-CoV-2 Mpro at 1.93 A resolution-5 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Ertem, B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK6
 
 | |
7VJZ
 
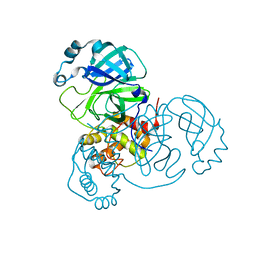 | | Crystal Structure of SARS-CoV-2 Mpro at 1.90 A resolution-7 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Tokay, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK7
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.4 A resolution-11 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Dag, C. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK2
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.0 A resolution -9 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Gul, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK8
 
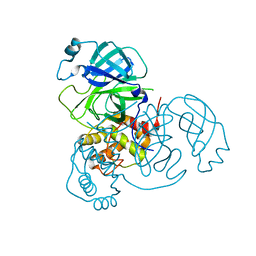 | | Crystal structure of SARS-CoV-2 Mpro at 2.4 A Resolution | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Usta, G. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VJX
 
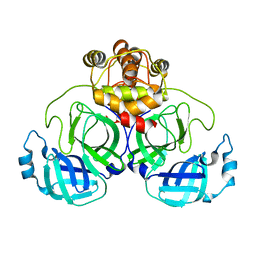 | | Crystal Structure of SARS-CoV-2 Mpro at 2.20 A resolution-12 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Usta, G. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VK4
 
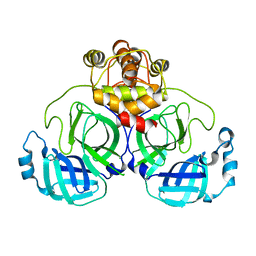 | |
7VJY
 
 | |
