1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
1UHR
 
 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
1UHW
 
 | | Solution structure of the DEP domain of mouse pleckstrin | | Descriptor: | Pleckstrin | | Authors: | Inoue, K, Yoshida, M, Hatta, R, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-11 | | Release date: | 2004-01-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DEP domain of mouse pleckstrin
To be Published
|
|
1UHZ
 
 | | Solution structure of dsRNA binding domain in Staufen homolog 2 | | Descriptor: | staufen (RNA binding protein) homolog 2 | | Authors: | He, F, Muto, Y, Obayashi, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Koboyashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dsRNA binding domain in Staufen homolog 2
To be Published
|
|
1UIL
 
 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | Descriptor: | Double-stranded RNA-binding motif | | Authors: | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
1WRB
 
 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | Descriptor: | DjVLGB, SULFATE ION | | Authors: | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
7T71
 
 | | Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus | | Descriptor: | Mevalonate 3,5-bisphosphate decarboxylase, OLEIC ACID | | Authors: | Vinokur, J.M, Sawaya, M.R, Cascio, D, Collazo, M, Bowie, J.U. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways.
J.Biol.Chem., 298, 2022
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8UH8
 
 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8UH5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8UH9
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
4NJV
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | Descriptor: | Protease, RITONAVIR | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJU
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with tipranavir | | Descriptor: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJT
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U8W
 
 | | HIV-1 wild Type protease with GRL-050-10A (a Gem-difluoro-bis-Tetrahydrofuran as P2-Ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of gem-Difluoro-bis-Tetrahydrofuran as P2 Ligand for HIV-1 Protease Inhibitors to Improve Brain Penetration: Synthesis, X-ray Studies, and Biological Evaluation.
Chemmedchem, 10, 2015
|
|
1N27
 
 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
3OK9
 
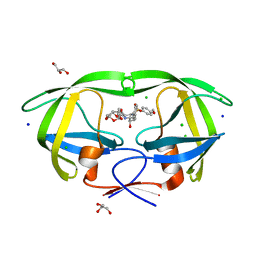 | | Crystal structure of wild-type HIV-1 protease with new oxatricyclic designed inhibitor GRL-0519A | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Probing Multidrug-Resistance and Protein-Ligand Interactions with Oxatricyclic Designed Ligands in HIV-1 Protease Inhibitors.
Chemmedchem, 5, 2010
|
|
7TO5
 
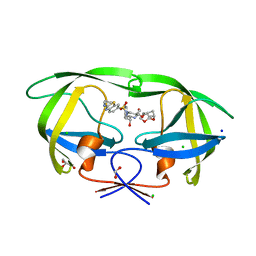 | | HIV-1 wild type protease with GRL-05816A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 1] | | Descriptor: | (1R,3aS,4S,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
7TO6
 
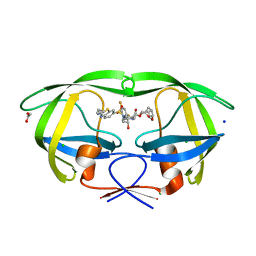 | | HIV-1 wild type protease with GRL-01717A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 2] | | Descriptor: | (1S,3aR,4R,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
2RPP
 
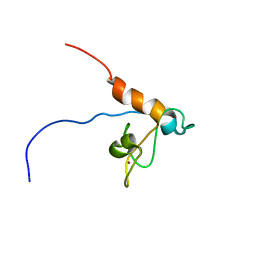 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
2RR4
 
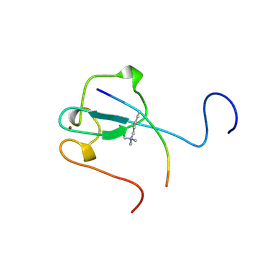 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
6CDL
 
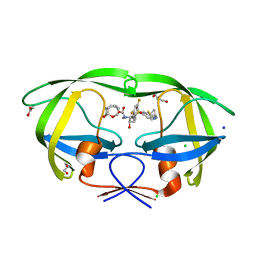 | | HIV-1 wild type protease with GRL-03214A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aR,4S,4aR,7aR,7bR)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
