2XZ6
 
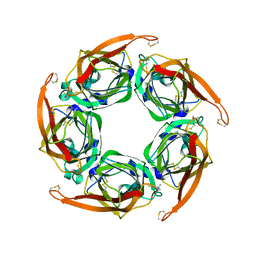 | | MTSET-modified Y53C mutant of Aplysia AChBP | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Gay, E.A, Colon Saez, J, Guskov, A, Van Elk, R, Van Der Schors, R.C, Peigneur, S, Tytgat, J, Strelkov, S.V, Smit, A.B, Yakel, J.L, Ulens, C. | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.137 Å) | | Cite: | Crystal Structures of a Cysteine-Modified Mutant in Loop D of Acetylcholine Binding Protein
J.Biol.Chem., 286, 2011
|
|
2XYS
 
 | | Crystal structure of Aplysia californica AChBP in complex with strychnine | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, STRYCHNINE | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2XYT
 
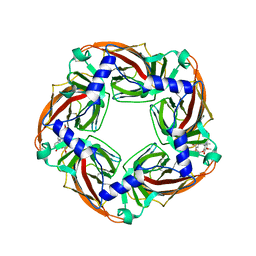 | | Crystal structure of Aplysia californica AChBP in complex with d- tubocurarine | | Descriptor: | D-TUBOCURARINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2YME
 
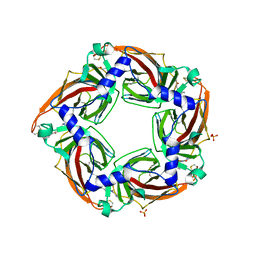 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
2MD6
 
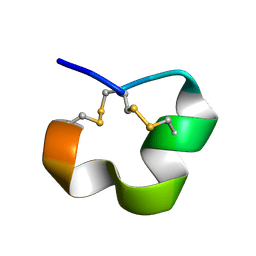 | | NMR SOLUTION STRUCTURE OF ALPHA CONOTOXIN LO1A FROM Conus longurionis | | Descriptor: | ALPHA CONOTOXIN LO1A | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Lebbe, E.K.M, Peigneur, S, D'Souza, L, Tytgat, J. | | Deposit date: | 2013-09-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Elucidation of a New alpha-Conotoxin, Lo1a, from Conus longurionis.
J.Biol.Chem., 289, 2014
|
|
2MOA
 
 | |
