4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
2D4Y
 
 | |
2D4X
 
 | |
2DPY
 
 | |
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
8YYM
 
 | | Cryo-EM structure of cylindrical fiber of MyD88 TIR | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Kasai, K, Imamura, K, Narita, A, Makino, F, Miyata, T, Kato, T, Namba, K, Onishi, H, Tochio, H. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | From Monomers to Oligomers: Structural Mechanism of Receptor-Triggered MyD88 Assembly in Innate Immune Signaling
Biorxiv, 2024
|
|
7VW6
 
 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
7W2J
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7WSQ
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
6JZT
 
 | |
6JF2
 
 | |
6JZR
 
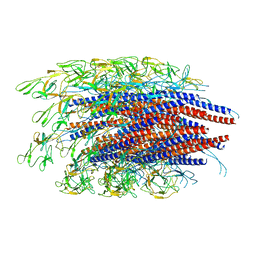 | |
7CLR
 
 | | CryoEM structure of S.typhimurium flagellar LP ring | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Yamaguchi, T, Makino, F, Miyata, T, Minamino, T, Kato, T, Namba, K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-02 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the molecular bushing of the bacterial flagellar motor.
Nat Commun, 12, 2021
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7EHA
 
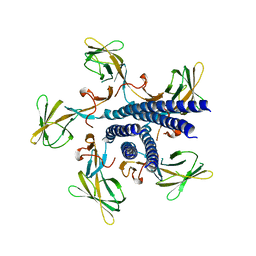 | | Crystal structure of the flagellar hook cap from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
7EH9
 
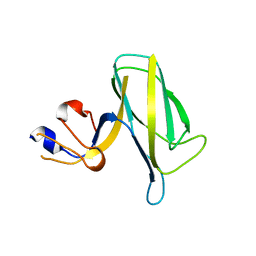 | | Crystal structure of the flagellar hook cap fragment from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
2E0Z
 
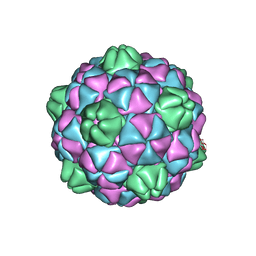 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
2ZVY
 
 | |
2ZF8
 
 | | Crystal structure of MotY | | Descriptor: | Component of sodium-driven polar flagellar motor | | Authors: | Imada, K, Kojima, S, Namba, K, Homma, M. | | Deposit date: | 2007-12-25 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZOV
 
 | |
3A7M
 
 | | Structure of FliT, the flagellar type III chaperone for FliD | | Descriptor: | Flagellar protein fliT | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7BWM
 
 | |
2ZVZ
 
 | |
3AJC
 
 | | Structure of the MC domain of FliG (PEV), a CW-biased mutant | | Descriptor: | Flagellar motor switch protein fliG | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rotational switching mechanism of the bacterial flagellar motor
Plos Biol., 9, 2011
|
|
6JY0
 
 | | CryoEM structure of S.typhimurium R-type straight flagellar filament made of FljB (A461V) | | Descriptor: | Flagellin | | Authors: | Yamaguchi, T, Toma, S, Terahara, N, Miyata, T, Minamino, T, Ashikara, M, Namba, K, Kato, T. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural and Functional Comparison ofSalmonellaFlagellar Filaments Composed of FljB and FliC.
Biomolecules, 10, 2020
|
|
