1KQU
 
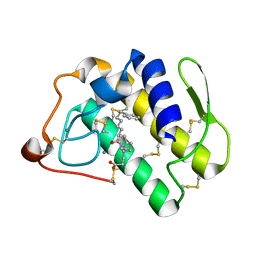 | | Human phospholipase A2 complexed with a substrate anologue | | Descriptor: | 6-PHENYL-4(R)-(7-PHENYL-HEPTANOYLAMINO)-HEXANOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Tyndall, J.D, Martin, J.L. | | Deposit date: | 2002-01-07 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D-Tyrosine as a chiral precusor to potent inhibitors of human nonpancreatic secretory phospholipase A2 (IIa) with antiinflammatory activity.
Chembiochem, 4, 2003
|
|
3PUK
 
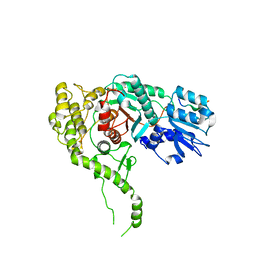 | | Re-refinement of the crystal structure of Munc18-3 and Syntaxin4 N-peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 3 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4ZL7
 
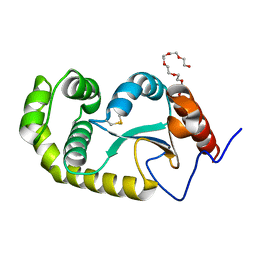 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal I | | Descriptor: | HEXAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL8
 
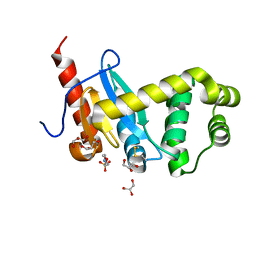 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahoh, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL9
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal III | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5ID4
 
 | |
5IDR
 
 | |
2Y92
 
 | | Crystal structure of MAL adaptor protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TOLL/INTERLEUKIN-1 RECEPTOR DOMAIN-CONTAINING ADAPTER PROTEIN, | | Authors: | Valkov, E, Stamp, A, Martin, J.L, Kobe, B. | | Deposit date: | 2011-02-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Toll-Like Receptor Adaptor Mal/Tirap Reveals the Molecular Basis for Signal Transduction and Disease Protection.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BCK
 
 | |
5DCH
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I in complex with MIPS-0000851 (3-[(2-METHYLBENZYL)SULFANYL]-4H-1,2,4-TRIAZOL-4-AMINE) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, GLYCEROL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
3BD2
 
 | |
5KBC
 
 | |
6MHH
 
 | | Proteus mirabilis ScsC linker (residues 39-49) deletion and N6K mutant | | Descriptor: | Metal resistance protein | | Authors: | Furlong, E.J, Martin, J.L. | | Deposit date: | 2018-09-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1N7J
 
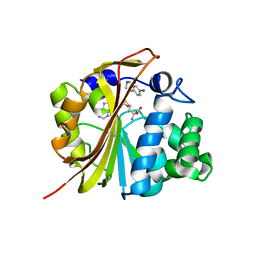 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
6NEN
 
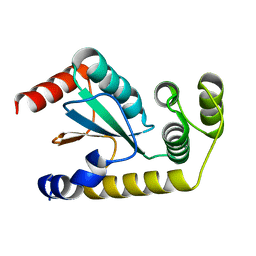 | | Catalytic domain of Proteus mirabilis ScsC | | Descriptor: | Copper resistance protein | | Authors: | Kurth, F, Furlong, E.J, Premkumar, L, Martin, J.L. | | Deposit date: | 2018-12-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1N7I
 
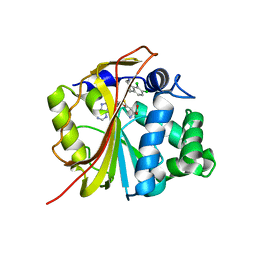 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1LS6
 
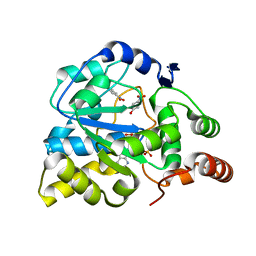 | | Human SULT1A1 complexed with PAP and p-Nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, aryl sulfotransferase | | Authors: | Gamage, N.U, Barnett, A.C, Tresillian, M, Latham, C.F, Liyou, N.E, McManus, M.E, Martin, J.L. | | Deposit date: | 2002-05-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition.
J.Biol.Chem., 278, 2003
|
|
1NOT
 
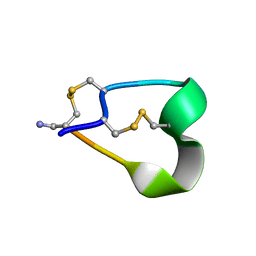 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
1MTR
 
 | | HIV-1 PROTEASE COMPLEXED WITH A CYCLIC PHE-ILE-VAL PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Wickramasinghe, W, Begun, J, Martin, J.L. | | Deposit date: | 1996-02-15 | | Release date: | 1996-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-based cyclic peptidomimetics of Phe-Ile-Val that inhibit HIV-1 protease using a novel enzyme-binding mode.
J.Am.Chem.Soc., 118, 1996
|
|
3KR0
 
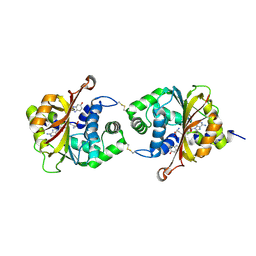 | |
3KPU
 
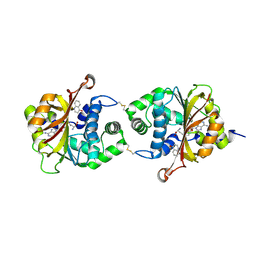 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-quinolinol | | Descriptor: | Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, quinolin-4-ol | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KR2
 
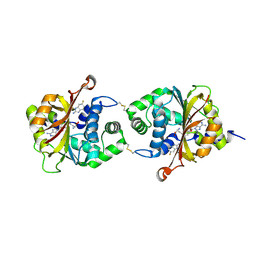 | |
3KQM
 
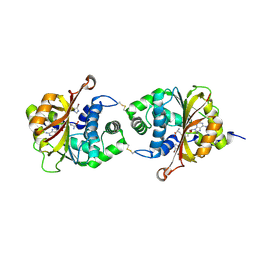 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-Bromo-1H-imidazole | | Descriptor: | 4-bromo-1H-imidazole, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQS
 
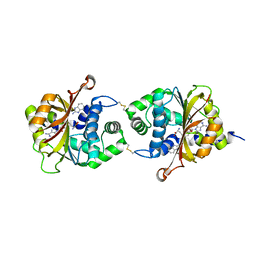 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Aminobenzimidazole | | Descriptor: | 1H-benzimidazol-2-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQY
 
 | |
