2MUY
 
 | | The solution structure of the FtsH periplasmic N-domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
2MV3
 
 | | The N-domain of the AAA metalloproteinase Yme1 from Saccharomyces cerevisiae | | Descriptor: | Mitochondrial inner membrane i-AAA protease supercomplex subunit YME1 | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
2LFS
 
 | | Solution structure of the chimeric Af1503 HAMP- EnvZ DHp homodimer; A219F variant | | Descriptor: | HAMP domain-containing protein, Osmolarity sensor protein EnzV chimera | | Authors: | Coles, M, Ferris, H.U, Hulko, M, Martin, J, Lupas, A.N. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of regulation of receptor histidine kinases.
Structure, 20, 2012
|
|
3WQA
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1ii) | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPP
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1iii) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPR
 
 | | Acinetobacter sp. Tol 5 AtaA N-terminal half of C-terminal stalk fused to GCN4 adaptors (CstalkN) | | Descriptor: | Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3ZRW
 
 | | The structure of the dimeric Hamp-Dhp fusion A291V mutant | | Descriptor: | AF1503 PROTEIN, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J, Lupas, A.N. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
5A1Q
 
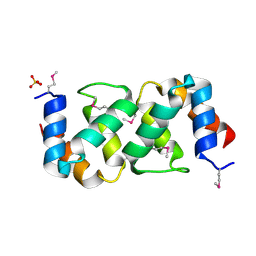 | |
5APW
 
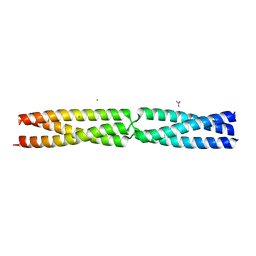 | | Sequence MATKDD inserted between GCN4 adaptors - Structure T6 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, ... | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5AMK
 
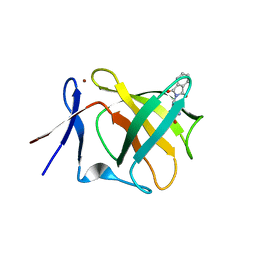 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in multiple conformations, hexagonal crystal form | | Descriptor: | CEREBLON ISOFORM 4, DIMETHYL SULFOXIDE, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5APX
 
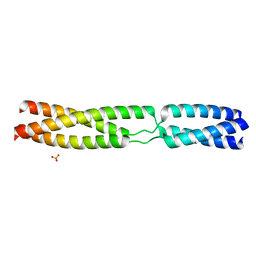 | | Sequence MATKDDIAN inserted between GCN4 adaptors - Structure T9(6) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PHOSPHATE ION | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APT
 
 | |
5APS
 
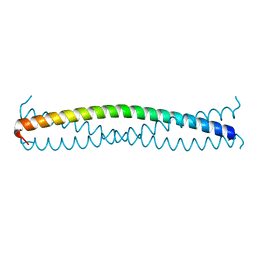 | |
5APZ
 
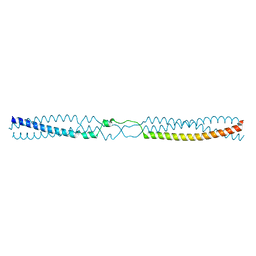 | | Thermosinus carboxydivorans Nor1 Tcar0761 residues 68-101 and 191-211 fused to GCN4 adaptors | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, NOR1 TCAR0761 | | Authors: | Hartmann, M.D, Deiss, S, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APP
 
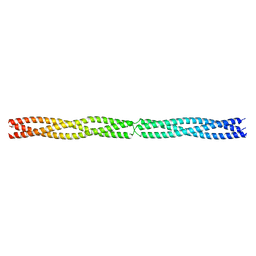 | | Actinobacillus actinomycetemcomitans OMP100 residues 133-198 fused to GCN4 adaptors | | Descriptor: | CHLORIDE ION, General control protein GCN4,GENERAL CONTROL PROTEIN GCN4, OUTER MEMBRANE PROTEIN 100,General control protein GCN4 | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APV
 
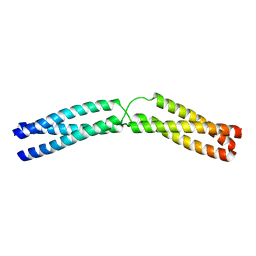 | |
5APY
 
 | |
5AMJ
 
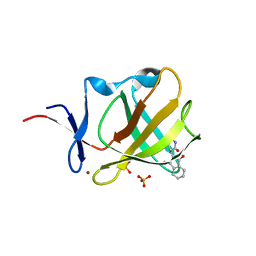 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash II structure | | Descriptor: | CEREBLON ISOFORM 4, PHOSPHATE ION, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5APU
 
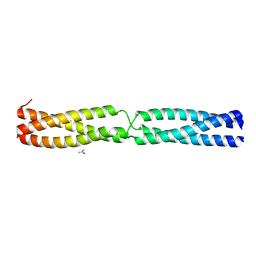 | | Sequence IANKEDKAD inserted between GCN4 adaptors - Structure A9b black | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, UREA | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APQ
 
 | |
5AMI
 
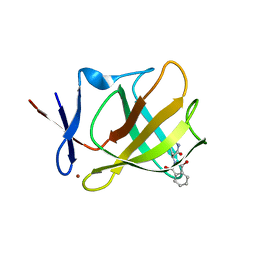 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash I structure | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5AMH
 
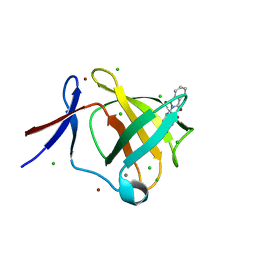 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, trigonal crystal form | | Descriptor: | CALCIUM ION, CEREBLON ISOFORM 4, CHLORIDE ION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
2W1T
 
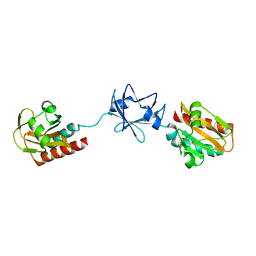 | | Crystal Structure of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WQ1
 
 | |
