5J58
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 1856) | | Descriptor: | (3S)-1-(5-chloro-1H-imidazo[4,5-b]pyridin-2-yl)-N-[(3,5-dichlorophenyl)methyl]piperidin-3-amine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-04-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided design of novel Trypanosoma brucei Methionyl-tRNA synthetase inhibitors.
Eur J Med Chem, 124, 2016
|
|
3EZJ
 
 | | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody | | Descriptor: | CHLORIDE ION, General secretion pathway protein GspD, NANOBODY NBGSPD_7, ... | | Authors: | Korotkov, K.V, Pardon, E, Steyaert, J, Hol, W.G. | | Deposit date: | 2008-10-22 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
Structure, 17, 2009
|
|
3LAD
 
 | |
2SBT
 
 | | A COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF SUBTILISIN BPN AND SUBTILISIN NOVO | | Descriptor: | ACETONE, SUBTILISIN NOVO | | Authors: | Drenth, J, Hol, W.G.J, Jansonius, J.N, Koekoek, R. | | Deposit date: | 1976-09-07 | | Release date: | 1976-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo.
Cold Spring Harbor Symp.Quant.Biol., 36, 1972
|
|
5TIM
 
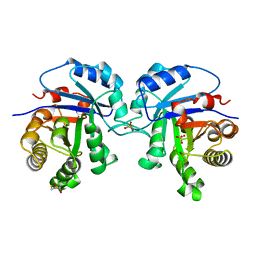 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
3SOL
 
 | |
3TGH
 
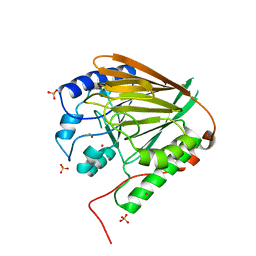 | | GAP50 the anchor in the inner membrane complex of Plasmodium | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Glideosome-associated protein 50, ... | | Authors: | Bosch, J, Paige, M.H, Vaidya, A, Bergman, L, Hol, W.G.J. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of GAP50, the anchor of the invasion machinery in the inner membrane complex of Plasmodium falciparum.
J.Struct.Biol., 178, 2012
|
|
3G20
 
 | | Crystal structure of the major pseudopilin from the type 2 secretion system of enterohaemorrhagic Escherichia coli | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Korotkov, K.V, Gray, M.D, Kreger, A, Turley, S, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Calcium is essential for the major pseudopilin in the type 2 secretion system.
J.Biol.Chem., 284, 2009
|
|
3FU1
 
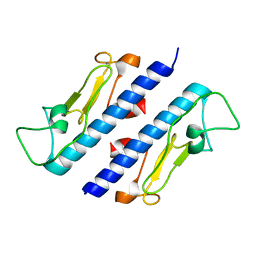 | | Crystal structure of the major pseudopilin from the type 2 secretion system of Vibrio cholerae | | Descriptor: | CALCIUM ION, General secretion pathway protein G, ZINC ION | | Authors: | Korotkov, K.V, Gray, M.D, Kreger, A, Turley, S, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium is essential for the major pseudopilin in the type 2 secretion system.
J.Biol.Chem., 284, 2009
|
|
3GN9
 
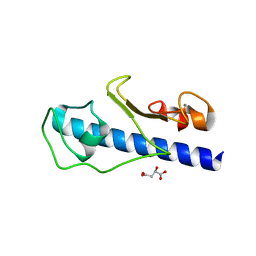 | | Crystal structure of the major pseudopilin from the type 2 secretion system of Vibrio vulnificus | | Descriptor: | CALCIUM ION, D-MALATE, Type II secretory pathway, ... | | Authors: | Korotkov, K.V, Gray, M.D, Kreger, A, Turley, S, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2009-03-16 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Calcium is essential for the major pseudopilin in the type 2 secretion system.
J.Biol.Chem., 284, 2009
|
|
2QV8
 
 | |
2RET
 
 | | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the Type 2 Secretion System of Vibrio vulnificus | | Descriptor: | CHLORIDE ION, EpsJ, Pseudopilin EpsI, ... | | Authors: | Yanez, M.E, Korotkov, K.V, Abendroth, J, Hol, W.G.J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
J.Mol.Biol., 375, 2008
|
|
2QAC
 
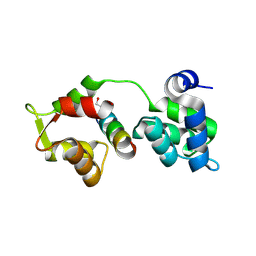 | | The closed MTIP-MyosinA-tail complex from the malaria parasite invasion machinery | | Descriptor: | Myosin A tail domain interacting protein MTIP, Myosin-A | | Authors: | Bosch, J, Turley, S, Roach, C.M, Daly, T.M, Bergman, L.W, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-06-14 | | Release date: | 2007-06-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Closed MTIP-Myosin A-Tail Complex from the Malaria Parasite Invasion Machinery.
J.Mol.Biol., 372, 2007
|
|
2TEC
 
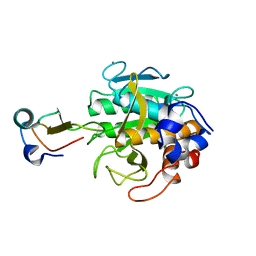 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
2PC4
 
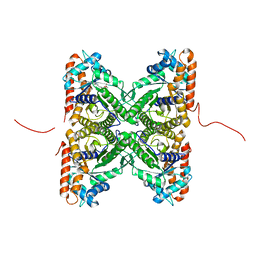 | | Crystal structure of fructose-bisphosphate aldolase from Plasmodium falciparum in complex with TRAP-tail determined at 2.4 angstrom resolution | | Descriptor: | Fructose-bisphosphate aldolase, PbTRAP | | Authors: | Bosch, J, Buscaglia, C.A, Krumm, B, Cardozo, T, Nussenzweig, V, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2VMB
 
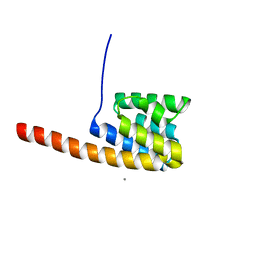 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
1HCY
 
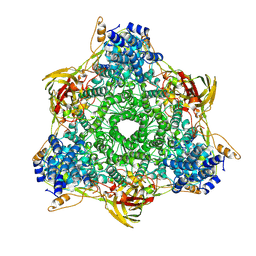 | |
4MW2
 
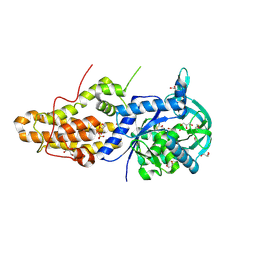 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-2-hydroxy-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1472) | | Descriptor: | 1-(3-{[5-chloro-2-hydroxy-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW6
 
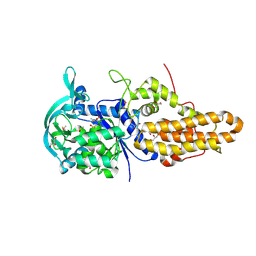 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[2-(benzyloxy)-5-chloro-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1476) | | Descriptor: | 1-(3-{[2-(benzyloxy)-5-chloro-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW7
 
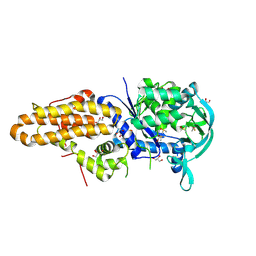 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1469) | | Descriptor: | 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW0
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (Chem 1392) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW5
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-phenylurea (Chem 1415) | | Descriptor: | 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-phenylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MVW
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MWE
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1475) | | Descriptor: | 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW4
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(5-chloro-2-hydroxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1473) | | Descriptor: | 1-{3-[(5-chloro-2-hydroxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
