5BP8
 
 | | ent-Copalyl diphosphate synthase from Streptomyces platensis | | Descriptor: | 1,2-ETHANEDIOL, Ent-copalyl diphosphate synthase, SULFATE ION | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Ma, M, Chang, C.-Y, Shen, B, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
1ZX5
 
 | | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-06 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus
To be Published
|
|
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
1ZX3
 
 | | Structure of NE0241 Protein of Unknown Function from Nitrosomonas europaea | | Descriptor: | hypothetical protein NE0241 | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of hypothetical protein NE0241 from Nitrosomonas europaea.
To be Published
|
|
1ZS7
 
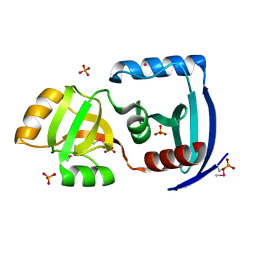 | | The structure of gene product APE0525 from Aeropyrum pernix | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein APE0525 | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of gene product APE0525 from Aeropyrum pernix
To be Published
|
|
5DA8
 
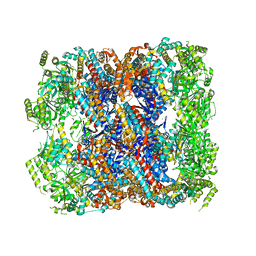 | | Crystal structure of chaperonin GroEL from | | Descriptor: | 60 kDa chaperonin, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Marshall, N, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chaperonin GroEL from
To Be Published
|
|
1XVI
 
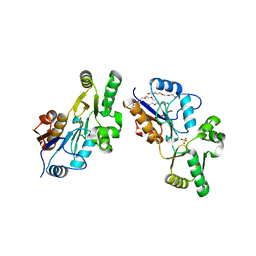 | | Crystal Structure of YedP, phosphatase-like domain protein from Escherichia coli K12 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative mannosyl-3-phosphoglycerate phosphatase, SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, A, Cymborowski, M, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of YedP, phosphatase-like domain protein from Escherichia coli K12
To be Published
|
|
1XPP
 
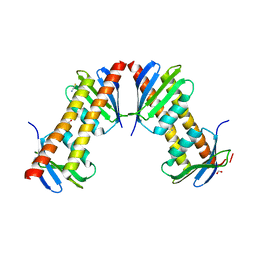 | | Crystal Structure of TA1416,DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum | | Descriptor: | ACETIC ACID, DNA-directed RNA polymerase subunit L, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of TA1416, DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum
To be Published
|
|
5CR9
 
 | | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017 | | Descriptor: | ABC-type Fe3+-hydroxamate transport system, periplasmic component, GLUTAMIC ACID, ... | | Authors: | Nocek, B, Cuff, M, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017
To Be Published
|
|
2A9S
 
 | | The crystal structure of competence/damage inducible protein CihA from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, competence/damage-inducible protein CinA | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Kim, Y, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of competence/damage inducible protein CihA from Agrobacterium tumefaciens
To be Published
|
|
2A61
 
 | | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima | | Descriptor: | transcriptional regulator Tm0710 | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Chang, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima
To be Published
|
|
2A35
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA4017 from Pseudomonas aeruginosa PAO1, Possible Epimerase | | Descriptor: | hypothetical protein PA4017 | | Authors: | Zhang, R, Xu, L, Cuff, M, Savchenko, A, Cymborowski, M, Minor, W, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-08-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA4017 from
Pseudomonas aeruginosa PAO1
To be Published
|
|
2FCK
 
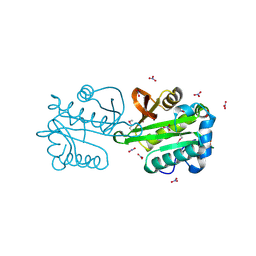 | | Structure of a putative ribosomal-protein-serine acetyltransferase from Vibrio cholerae. | | Descriptor: | GLYCEROL, NITRATE ION, ribosomal-protein-serine acetyltransferase, ... | | Authors: | Cuff, M.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-28 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an acetyltransferase protein from Vibrio cholerae strain N16961.
Proteins, 69, 2007
|
|
4DU6
 
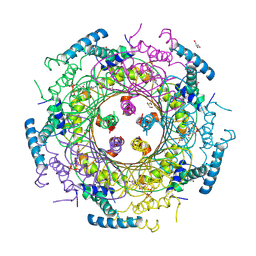 | | Crystal structure of GTP cyclohydrolase I from Yersinia pestis complexed with GTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Crystal structure of GTP cyclohydrolase I from Yersinia pestis complexed with GTP
To be Published
|
|
4DQD
 
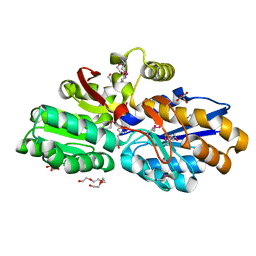 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4DZR
 
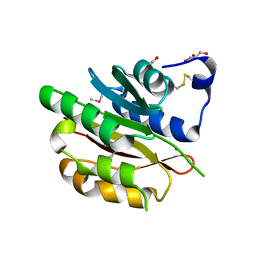 | | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
2IQY
 
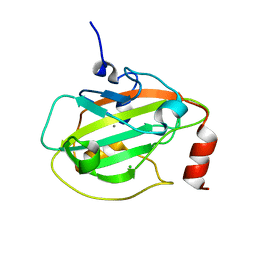 | | Rat Phosphatidylethanolamine-Binding Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Heil, G.L, Koide, S, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Rat Phosphatidylethanolamine-Binding Protein
To be Published
|
|
4DIB
 
 | | The crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Bacillus anthracis str. Sterne | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | Authors: | Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Bacillus anthracis str. Sterne
To be Published
|
|
4DQ1
 
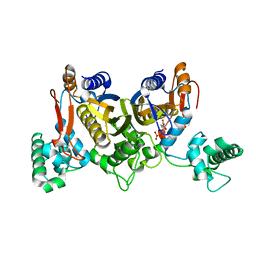 | | Thymidylate synthase from Staphylococcus aureus. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Osipiuk, J, Holowicki, J, Jedrzejczak, R, Rubin, E, Guinn, K, Ioerger, T, Baker, D, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-14 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Thymidylate synthase from Staphylococcus aureus.
To be Published
|
|
4DQ0
 
 | |
4DYU
 
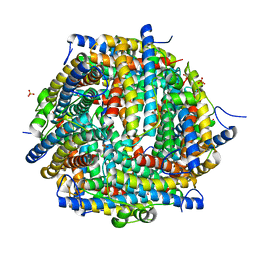 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4E16
 
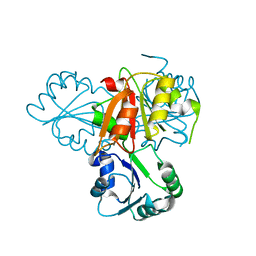 | | Precorrin-4 C(11)-methyltransferase from Clostridium difficile | | Descriptor: | precorrin-4 C(11)-methyltransferase | | Authors: | Osipiuk, J, Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-05 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Precorrin-4 C(11)-methyltransferase from Clostridium difficile
To be Published
|
|
4DWJ
 
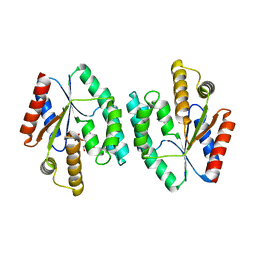 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate | | Descriptor: | THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate
To be Published
|
|
2IQX
 
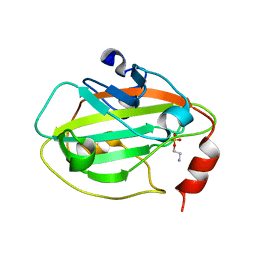 | | Rat Phosphatidylethanolamine-Binding Protein Containing the S153E Mutation in the Complex with o-Phosphorylethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Clark, M.C, Rosner, M, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rat Phosphatidylethanolamine-Binding Crystal structure of Protein Containing the S153E Mutation in the Complex with o-Phosphorylethanolamine
To be Published
|
|
4E2G
 
 | | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Cupin 2 conserved barrel domain protein, NICKEL (II) ION, ... | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-08 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus
To be Published
|
|
