7O1C
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome-RF2 complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1A
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
8RSA
 
 | |
7O0M
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 9 (ADO_AD_023) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-[6-[(phenylmethyl)amino]pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 9 (ADO_AD_023)
To Be Published
|
|
7O28
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 19 (ADO_AE_009) | | Descriptor: | 9-(2-chloranyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 19 (ADO_AE_009)
To Be Published
|
|
7O2H
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 13 (ADO_AD_091) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-1-methyl-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 13 (ADO_AD_091)
To Be Published
|
|
7O0P
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 10 (ADO_AD_022) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 10 (ADO_AD_022)
To Be Published
|
|
7O0R
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 15 (ADO_AE_026) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 15 (ADO_AE_026)
To Be Published
|
|
7O27
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 17 (ADO_AE_005) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 17 (ADO_AE_005)
To Be Published
|
|
7O0Q
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 12 (ADO_AD_066) | | Descriptor: | 3-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-8-[6-(methylamino)pyrimidin-4-yl]-1,3,8-triazaspiro[4.5]decan-2-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 12 (ADO_AD_066)
To Be Published
|
|
7NR5
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-[(2-methyl-1,2,3-triazol-4-yl)amino]pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-(3-fluoranyl-5-methoxy-phenyl)-2-oxidanyl-ethyl]propanamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
7NR9
 
 | |
7NR8
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-2-oxidanyl-ethyl]propanamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | O'Reilly, M, Cleasby, A. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
3UMP
 
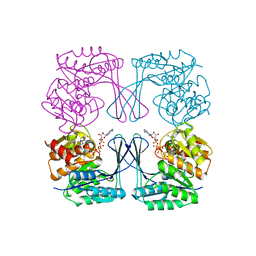 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with Cesium and ATP | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, CESIUM ION, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Garratt, R.C, Babul, J. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | A Ribokinase Family Conserved Monovalent Cation Binding Site Enhances the MgATP-induced Inhibition in E. coli Phosphofructokinase-2
Biophys.J., 105, 2013
|
|
7T88
 
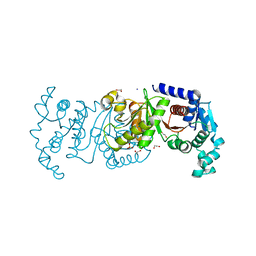 | | Crystal Structure of the C-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of c from Escherichia coli
To Be Published
|
|
9EM1
 
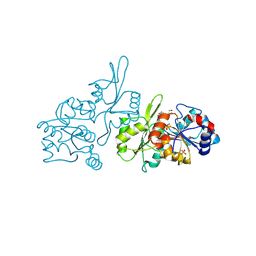 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
7T85
 
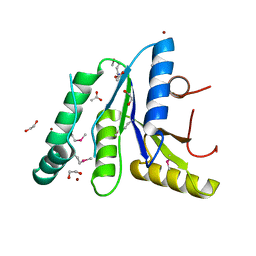 | | Crystal Structure of the N-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Phosphate acetyltransferase, ... | | Authors: | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the N-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli
To Be Published
|
|
7T3X
 
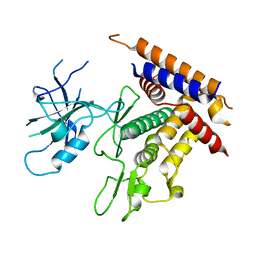 | | Structure of unphosphorylated Pediculus humanus (Ph) PINK1 D334A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1 | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4K
 
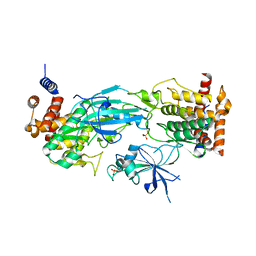 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with kinked alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4N
 
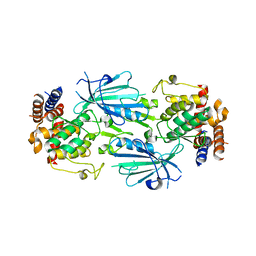 | | Structure of dimeric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T4L
 
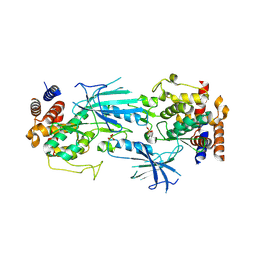 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
3URI
 
 | | Endothiapepsin-DB5 complex. | | Descriptor: | DB5 peptide, Endothiapepsin | | Authors: | Bailey, D, Sanz-Aparicio, J, Albert, A, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7T4M
 
 | | Structure of dodecameric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
3UMO
 
 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with Potassium | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Garratt, R.C, Babul, J. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | A Ribokinase Family Conserved Monovalent Cation Binding Site Enhances the MgATP-induced Inhibition in E. coli Phosphofructokinase-2
Biophys.J., 105, 2013
|
|
