7SWH
 
 | |
7SYC
 
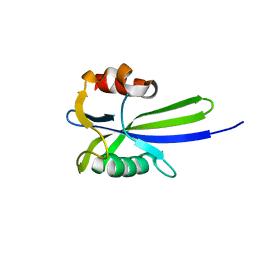 | |
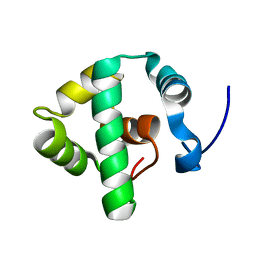
7SYB
 
 | |
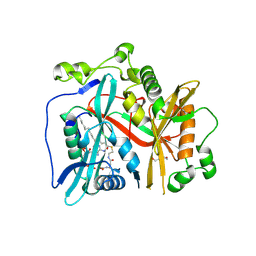
8FBT
 
 | |
8FBM
 
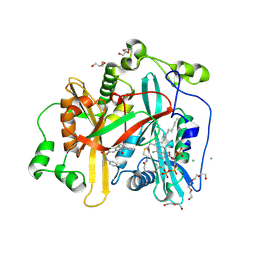 | |
7T35
 
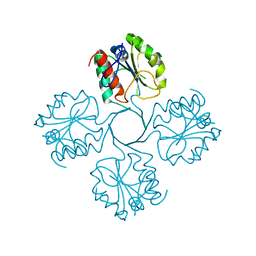 | |
7SY9
 
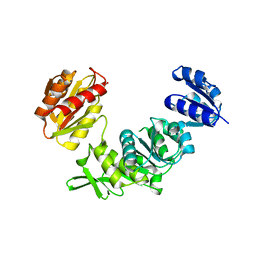 | |
8FBU
 
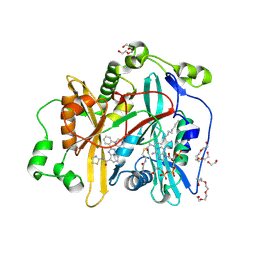 | |
7T24
 
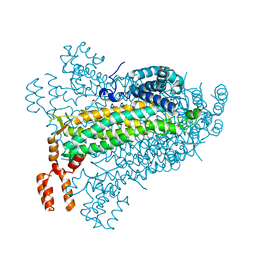 | |
7SZS
 
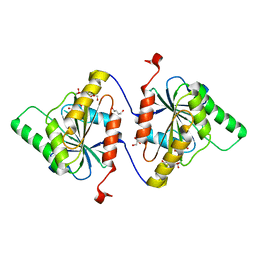 | |
7T29
 
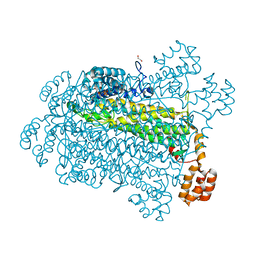 | |
7SZP
 
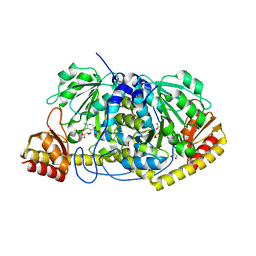 | |
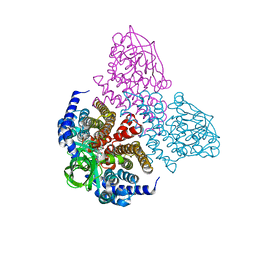
7SZV
 
 | |
7TAT
 
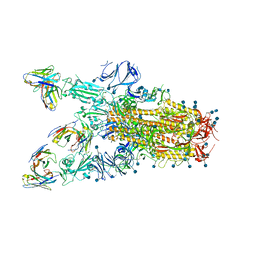 | |
7TAS
 
 | |
7TCM
 
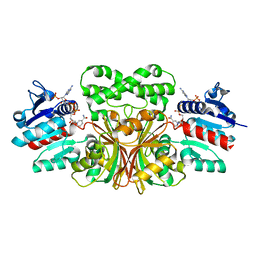 | |
8FXC
 
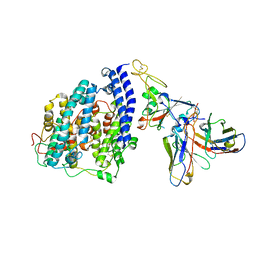 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7T7J
 
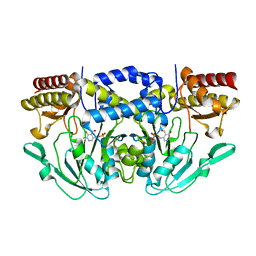 | |
7TA9
 
 | |
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7STZ
 
 | |
8FBQ
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 12b | | Descriptor: | 1-[(3M)-3-{3-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]pyridin-2-yl}phenyl]piperazine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Staker, B.L, Lovell, S.W, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of potent and selective N-myristoyltransferase inhibitors of Plasmodium vivax liver stage hypnozoites and schizonts.
Nat Commun, 14, 2023
|
|
3MEN
 
 | |
8FBP
 
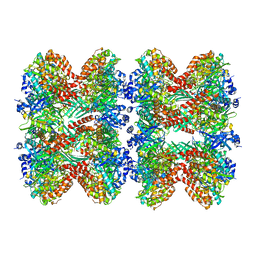 | | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation | | Descriptor: | Glutamine synthetase | | Authors: | Phan, I.Q, Staker, B, Shek, R, Moser, T.H, Evans, J.E, van Voorhis, W.C, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation
To Be Published
|
|
3NJB
 
 | |
