4C5D
 
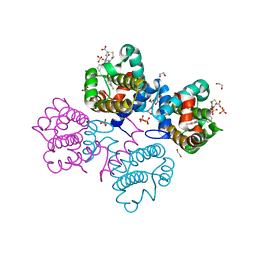 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (42) | | Descriptor: | (R)-3-(4-BROMOBENZYLTHIO)-2-(3-(3-((2,4-DIFLUOROPHENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
3ZK6
 
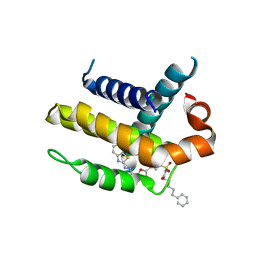 | | Crystal structure of Bcl-xL in complex with inhibitor (Compound 2). | | Descriptor: | BCL-2-LIKE PROTEIN 1, N-(3-(5-(1-(2-(benzo[d]thiazol-2-yl)hydrazono)ethyl)furan-2-yl)phenylsulfonyl)-6-phenylhexanamide | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
4C52
 
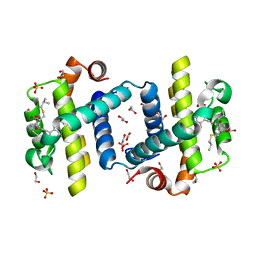 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
4CI2
 
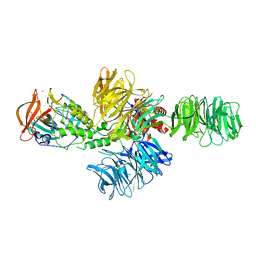 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4PZA
 
 | | The complex structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c with inorganic phosphate | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, PHOSPHATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-29 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4PZ9
 
 | | The native structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4QIH
 
 | | The structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c complexes with VO3 | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, VANADATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
8WCW
 
 | |
8WCX
 
 | |
8K11
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K12
 
 | | SID1 transmembrane family member 2 | | Descriptor: | SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1B
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K10
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K13
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1D
 
 | | SID1 transmembrane family member 1 | | Descriptor: | SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
5H2C
 
 | | Crystal structure of Saccharomyces cerevisiae Osh1 ANK - Nvj1 | | Descriptor: | Nucleus-vacuole junction protein 1, Oxysterol-binding protein homolog 1 | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2D
 
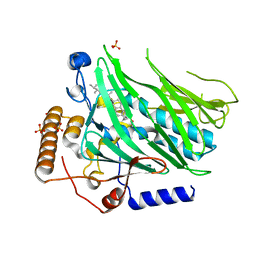 | | Crystal structure of Osh1 ORD domain in complex with ergosterol | | Descriptor: | ERGOSTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2A
 
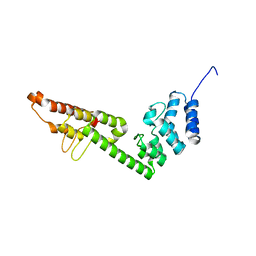 | |
5H28
 
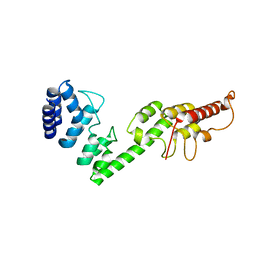 | |
8Y8C
 
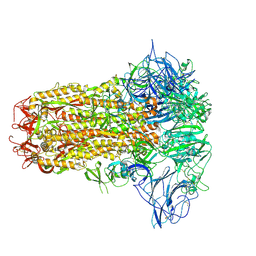 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8E
 
 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y7Y
 
 | | Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y, Liu, X, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y88
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y89
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
