3T5M
 
 | | Crystal structure of the S112A mutant of mycrocine immunity protein (MccF) with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
3VCX
 
 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | Descriptor: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
3TTG
 
 | | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, Putative aminomethyltransferase | | Authors: | Michalska, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum
TO BE PUBLISHED
|
|
1R8K
 
 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
3TYX
 
 | | Crystal structure of the F177S mutant of mycrocine immunity protein (MccF) with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin immunity protein MccF | | Authors: | Nocek, B, Gu, M, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
1L3L
 
 | | Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*GP*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP*CP*TP*GP*CP*AP*CP*AP*TP*C)-3', Transcriptional activator protein traR | | Authors: | Zhang, R, Pappas, T, Brace, J.L, Miller, P.C, Oulmassov, T, Molyneaux, J.M, Anderson, J.C, Bashkin, J.K, Winans, S.C, Joachimiak, A. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA.
Nature, 417, 2002
|
|
1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
6B8D
 
 | | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae.
To Be Published
|
|
6BBX
 
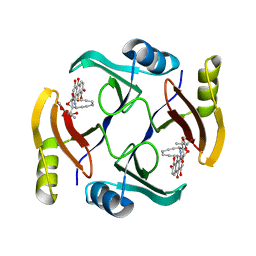 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
4R0J
 
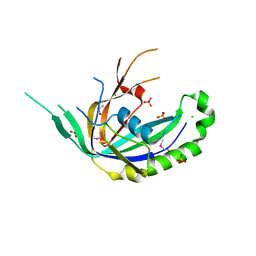 | | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans
To be Published
|
|
4RD8
 
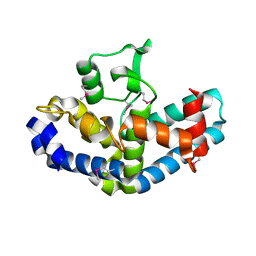 | | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4RHA
 
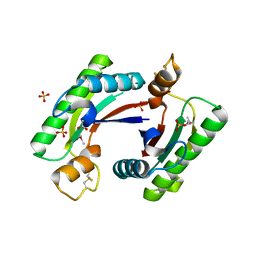 | | Structure of the C-terminal domain of outer-membrane protein OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Outer membrane protein A, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
6B7J
 
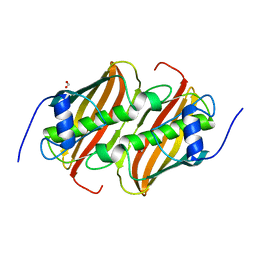 | |
6BLG
 
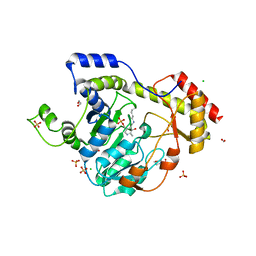 | | Crystal Structure of Sugar Transaminase from Klebsiella pneumoniae Complexed with PLP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal Structure of Sugar Transaminase from Klebsiella pneumoniae Complexed with PLP
To Be Published
|
|
6BMA
 
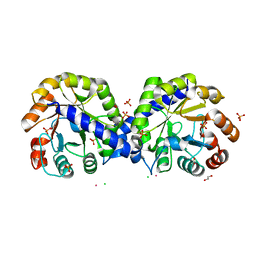 | | The crystal structure of indole-3-glycerol phosphate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Zhou, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-14 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
6BO0
 
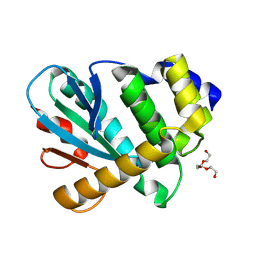 | | MdbA protein, a thiol-disulfide oxidoreductase from Corynebacterium matruchotii | | Descriptor: | MdbA protein, TETRAETHYLENE GLYCOL | | Authors: | Osipiuk, J, Luong, T.Y, Trigar, R, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of a Thiol-Disulfide Oxidoreductase in the Hedgehog-Forming Actinobacterium Corynebacterium matruchotii.
J. Bacteriol., 200, 2018
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
6CIA
 
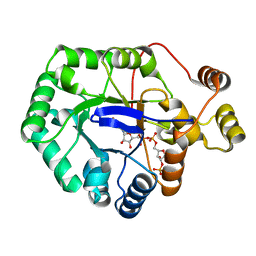 | | Crystal structure of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH. | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Leung, E.S, Shabalin, I.G, Grabowski, M, Almo, S.C, Satchell, K.J, Joachimiak, A, Lewinski, K, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH.
to be published
|
|
6CKY
 
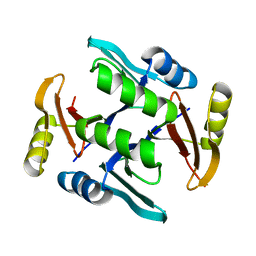 | | Crystal structure of UcmS2 | | Descriptor: | Glyoxalase | | Authors: | Chang, C.Y, Chang, C, Annaval, T, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UcmS2
To Be Published
|
|
6CMZ
 
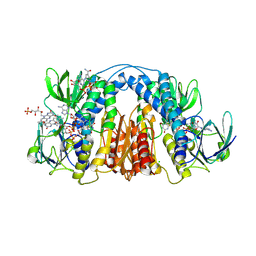 | | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D-MALATE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD.
To Be Published
|
|
6D0G
 
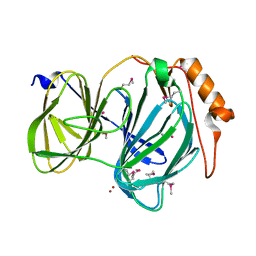 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
6DB1
 
 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6CA3
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
