8I04
 
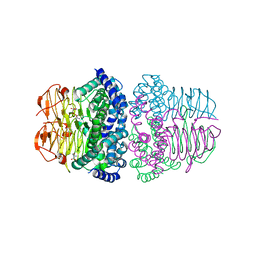 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
1EQ5
 
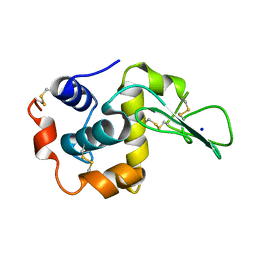 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQE
 
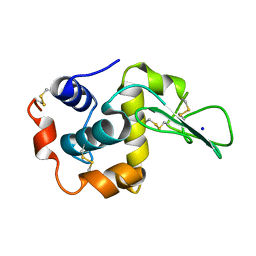 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-04 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
3A1M
 
 | | A fusion protein of a beta helix region of gene product 5 and the foldon region of bacteriophage T4 | | Descriptor: | POTASSIUM ION, chimera of thrombin cleavage site, Tail-associated lysozyme, ... | | Authors: | Yokoi, N, Suzuki, A, Hikage, T, Koshiyama, T, Terauchi, M, Yutani, K, Kanamaru, S, Arisaka, F, Yamane, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-04-11 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4
Small, 6, 2010
|
|
7YVW
 
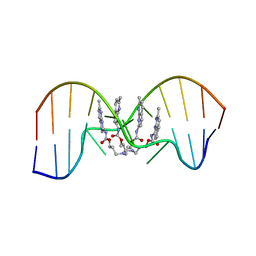 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
4TKC
 
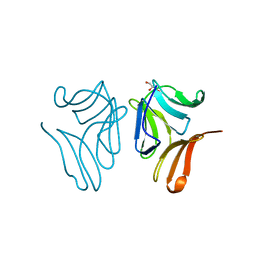 | | Japanese Marasmius oreades lectin complexed with mannose | | Descriptor: | GLYCEROL, Mannose recognizing lectin, alpha-D-mannopyranose, ... | | Authors: | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | Deposit date: | 2014-05-26 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of Japanese Marasmius oreades lectin complexed with mannose.
To Be Published
|
|
8WWD
 
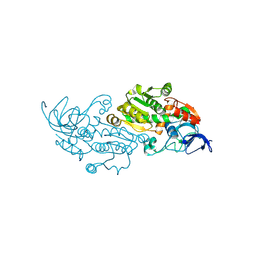 | |
8WWE
 
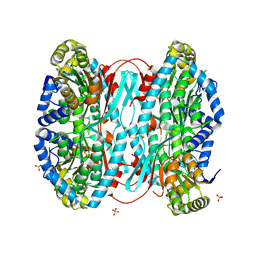 | |
8WWF
 
 | |
1EQ4
 
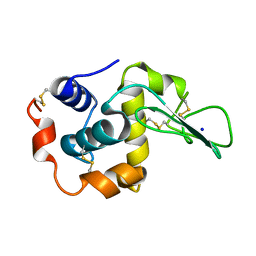 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
2H9V
 
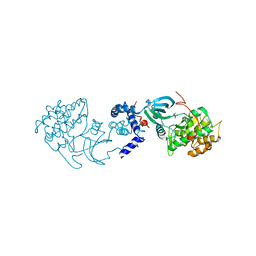 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1DI3
 
 | |
1DI4
 
 | |
1OUA
 
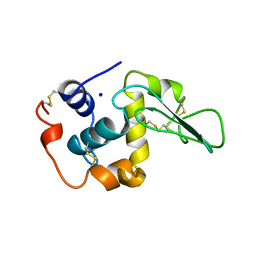 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE I56T MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure, stability, and folding process of amyloidogenic mutant human lysozyme.
J.Biochem.(Tokyo), 120, 1996
|
|
1WQO
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQM
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQR
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQP
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQN
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQQ
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
6QIS
 
 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model II) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3'), SULFATE ION | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
1GBY
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GB6
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GDX
 
 | |
