7S7D
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC SULFO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7RZD
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8E
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8J
 
 | | PHOSPHOPEPTIDE-SPECIFIC LC13 TCR, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, TRAV27_LC13 TCR ALPHA CHAIN, TRBV27_LC13 TCR BETA CHAIN | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S7E
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH DOT1L(998-1006) PEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S79
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC PHOSPHONO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8F
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8I
 
 | | PHOSPHOPEPTIDE-SPECIFIC LC13 TCR, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, TRAV27_LC13 TCR ALPHA CHAIN, TRBV27_LC13 TCR BETA CHAIN | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
3CFZ
 
 | |
1WEK
 
 | | Crystal structure of the conserved hypothetical protein TT1465 from Thermus thermophilus HB8 | | Descriptor: | PHOSPHATE ION, hypothetical protein TT1465 | | Authors: | Kukimoto-Niino, M, Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
7T62
 
 | | GPC2 HEP CT3 complex | | Descriptor: | CT3, Glypican-2 | | Authors: | Zhu, J, Cachau, R, De Val Alda, N, Li, N, Ho, M. | | Deposit date: | 2021-12-13 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice.
Cell Rep Med, 2, 2021
|
|
6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
1F11
 
 | | F124 FAB FRAGMENT FROM A MONOCLONAL ANTI-PRES2 ANTIBODY | | Descriptor: | F124 IMMUNOGLOBULIN (IGG1 HEAVY CHAIN), F124 IMMUNOGLOBULIN (KAPPA LIGHT CHAIN) | | Authors: | Saul, F.A, Vulliez-Le Normand, B, Passafiume, M, Riottot, M.M, Bentley, G.A. | | Deposit date: | 2000-05-18 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Fab fragment from F124, a monoclonal antibody specific for hepatitis B surface antigen.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1F4O
 
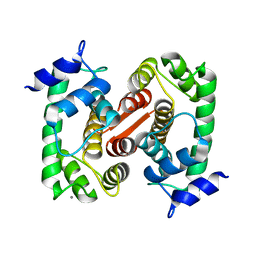 | | CRYSTAL STRUCTURE OF GRANCALCIN WITH BOUND CALCIUM | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2000-06-08 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
9EN6
 
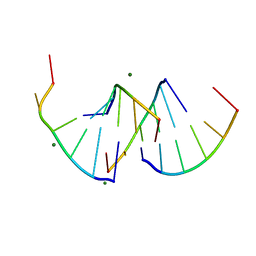 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Mateja-Pluta, M, Kiliszek, A. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.918 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 2024
|
|
6R3B
 
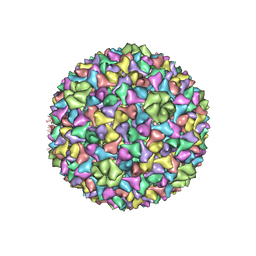 | | BACTERIOPHAGE SPP1 PROCAPSID-I PROTEIN | | Descriptor: | Major capsid protein | | Authors: | Ignatiou, A, Brasiles, S, El Sadek-Fadel, M, Buerger, J, Mielke, T, Topf, M, Tavares, P. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural transitions during the scaffolding-driven assembly of a viral capsid.
Nat Commun, 10, 2019
|
|
6R7U
 
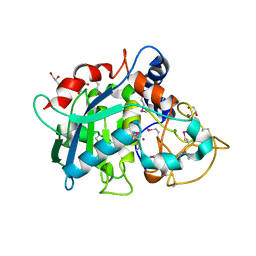 | | Selenomethionine variant of Tannerella forsythia promirolysin mutant E225A | | Descriptor: | BORIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
8HFP
 
 | |
6R7V
 
 | | Tannerella forsythia promirolysin mutant E225A | | Descriptor: | CALCIUM ION, GLYCEROL, Mirolysin, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
1FSS
 
 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH FASCICULIN-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, FASCICULIN II, ... | | Authors: | Harel, M, Kleywegt, G.J, Silman, I, Sussman, J.L. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an acetylcholinesterase-fasciculin complex: interaction of a three-fingered toxin from snake venom with its target.
Structure, 3, 1995
|
|
1FKT
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKR
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FP4
 
 | | CRYSTAL STRUCTURE OF THE ALPHA-H195Q MUTANT OF NITROGENASE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Sorlie, M, Christiansen, J, Lemon, B.J, Peters, J.W, Dean, D.R, Hales, B.J. | | Deposit date: | 2000-08-30 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic features and structure of the nitrogenase alpha-Gln195 MoFe protein
Biochemistry, 40, 2001
|
|
1WHF
 
 | | COAGULATION FACTOR, NMR, 15 STRUCTURES | | Descriptor: | COAGULATION FACTOR X | | Authors: | Sunnerhagen, M, Olah, G.A, Stenflo, J, Forsen, S, Drakenberg, T, Trewhella, J. | | Deposit date: | 1996-06-18 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The relative orientation of Gla and EGF domains in coagulation factor X is altered by Ca2+ binding to the first EGF domain. A combined NMR-small angle X-ray scattering study.
Biochemistry, 35, 1996
|
|
