4FIG
 
 | | Catalytic domain of human PAK4 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIJ
 
 | | Catalytic domain of human PAK4 | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6OMO
 
 | | Human BMP6 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 6, ... | | Authors: | Juo, Z.S, Seeherman, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix.
Sci Transl Med, 11, 2019
|
|
4FIE
 
 | | Full-length human PAK4 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2H2Z
 
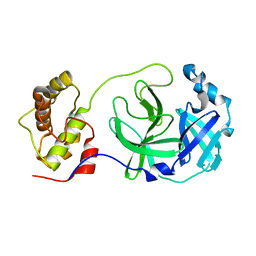 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
2JWY
 
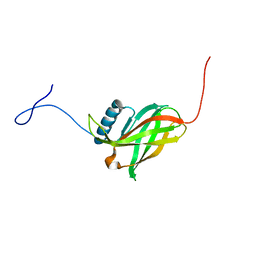 | | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli. Northeast Structural Genomics target ER540 | | Descriptor: | Uncharacterized lipoprotein yajI | | Authors: | Liu, G, Xiao, R, Chen, C, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli.
To be Published
|
|
6EET
 
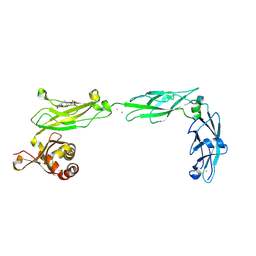 | | Crystal structure of mouse Protocadherin-15 EC9-MAD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Narui, Y, Sotomayor, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural determinants of protocadherin-15 mechanics and function in hearing and balance perception.
Proc.Natl.Acad.Sci.USA, 2020
|
|
6E8F
 
 | |
6EB5
 
 | |
3NL9
 
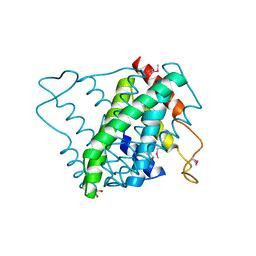 | |
3MX6
 
 | |
4HAC
 
 | |
2HOB
 
 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | Authors: | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
8UX6
 
 | | Structure of Fab201 with a T. parva sporozoite neutralizing B cell epitope of p67 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Singer, A.U, Gopalsamy, A, Fellouse, F.A, Miersch, S, Sidhu, S.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular characterization of a synthetic neutralizing antibody targeting p67 of Theileria parva.
Protein Sci., 34, 2025
|
|
5IGX
 
 | |
5I8D
 
 | |
8UHH
 
 | | anti-Phosphohistidine Fab hSC44.ck.elbow.20.N32F | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UIH
 
 | | anti-Phosphohistidine Fab hSC44.ck.20 with 3pTza peptide | | Descriptor: | 1,2-ETHANEDIOL, 3ptza peptide, hSC44.ck.20 Fab heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-10 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UHJ
 
 | | anti-Phosphohistidine Fab hSC44.ck.20.N32F | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, hSC44.ck.20.N32F Fab light chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UHP
 
 | | anti-Phosphohistidine Fab hSC44.ck.20.N32F with 3pTZA peptide | | Descriptor: | 1,2-ETHANEDIOL, 3pTza peptide, ACETATE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UIO
 
 | | anti-Phosphohistidine Fab hSC44.ck | | Descriptor: | CITRIC ACID, hSC44.ck Fab heavy chain, hSC44.ck Fab light chain | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-10 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UHT
 
 | |
8UHS
 
 | | anti-Phosphohistidine Fab hSC44.ck.20.elbow bound to phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, hSC44.ck.20.elbow Fab heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UHN
 
 | | anti-Phosphohistidine Fab hSC44.ck.20.N32F with 3pHis peptide | | Descriptor: | 1,2-ETHANEDIOL, 3pHis peptide, ACETATE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UIG
 
 | | anti-Phosphohistidine Fab hSC44.ck.20 with 3pHis peptide | | Descriptor: | 3pHis peptide, hSC44.ck.20 Fab heavy chain, hSC44.ck.20 Fab light chain | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-10 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
