1NOJ
 
 | |
1NOK
 
 | |
1PYG
 
 | |
1PKM
 
 | |
1QRZ
 
 | | CATALYTIC DOMAIN OF PLASMINOGEN | | Descriptor: | PLASMINOGEN | | Authors: | Peisach, E, Wang, J, de los Santos, T, Reich, E, Ringe, D. | | Deposit date: | 1999-06-16 | | Release date: | 1999-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the proenzyme domain of plasminogen.
Biochemistry, 38, 1999
|
|
1DYR
 
 | | THE STRUCTURE OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE TO 1.9 ANGSTROMS RESOLUTION | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | Authors: | Champness, J.N, Achari, A, Ballantine, S.P, Bryant, P.K, Delves, C.J, Stammers, D.K. | | Deposit date: | 1994-09-14 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of Pneumocystis carinii dihydrofolate reductase to 1.9 A resolution.
Structure, 2, 1994
|
|
1GPY
 
 | |
1H04
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, NICKEL (II) ION | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H2P
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1HCF
 
 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | Authors: | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
2GPN
 
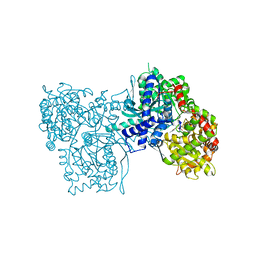 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
2Y0S
 
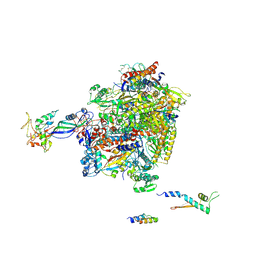 | | Crystal structure of Sulfolobus shibatae RNA polymerase in P21 space group | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, DNA-DIRECTED RNA POLYMERASE SUBUNIT A'', DNA-DIRECTED RNA POLYMERASE SUBUNIT D, ... | | Authors: | Wojtas, M, Peralta, B, Ondiviela, M, Mogni, M, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2010-12-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Archaeal RNA polymerase: the influence of the protruding stalk in crystal packing and preliminary biophysical analysis of the Rpo13 subunit.
Biochem. Soc. Trans., 39, 2011
|
|
4AYB
 
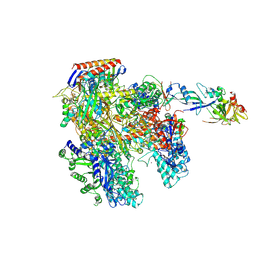 | | RNAP at 3.2Ang | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
2TNF
 
 | | 1.4 A RESOLUTION STRUCTURE OF MOUSE TUMOR NECROSIS FACTOR, TOWARDS MODULATION OF ITS SELECTIVITY AND TRIMERISATION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, PROTEIN (TUMOR NECROSIS FACTOR ALPHA) | | Authors: | Baeyens, K.J, De Bondt, H.L, Raeymaekers, A, Fiers, W, De Ranter, C.J. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of mouse tumour-necrosis factor at 1.4 A resolution: towards modulation of its selectivity and trimerization.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1A6S
 
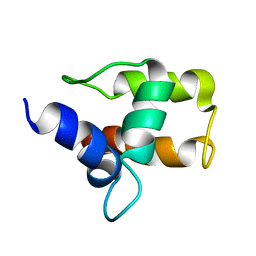 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
1A8I
 
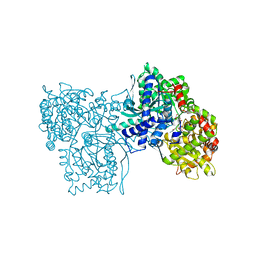 | | SPIROHYDANTOIN INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | BETA-D-GLUCOPYRANOSE SPIROHYDANTOIN, GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
1AXR
 
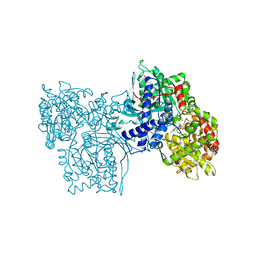 | | COOPERATIVITY BETWEEN HYDROGEN-BONDING AND CHARGE-DIPOLE INTERACTIONS IN THE INHIBITION OF BETA-GLYCOSIDASES BY AZOLOPYRIDINES: EVIDENCE FROM A STUDY WITH GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 4,5,6-TRIHYDROXY-7-HYDROXYMETHYL-4,5,6,7-TETRAHYDRO-1H-[1,2,3]TRIAZOLO[1,5-A]PYRIDIN-8-YLIUM, GLYCOGEN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Oikonomakos, N.G, Heightman, T.D. | | Deposit date: | 1997-10-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cooperative interactions of the catalytic nucleophile and the catalytic acid in the inhibition of beta-glycosidases. Calculations and their validation by comparative kinetic and structural studies of the inhibition of glycogen phosphorylase b.
HELV.CHIM.ACTA, 81, 1998
|
|
1BX3
 
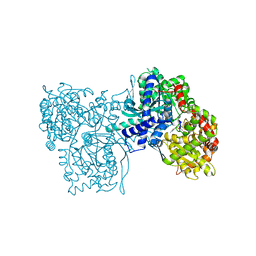 | | EFFECTS OF COMMONLY USED CRYOPROTECTANTS ON GLYCOGEN PHOSPHORYLASE ACTIVITY AND STRUCTURE | | Descriptor: | PROTEIN (GLYCOGEN PHOSPHORYLASE B), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1999-10-13 | | Release date: | 1999-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
1D63
 
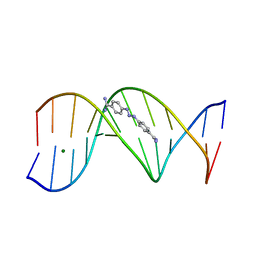 | | CRYSTAL STRUCTURE OF A BERENIL-D(CGCAAATTTGCG) COMPLEX; AN EXAMPLE OF DRUG-DNA RECOGNITION BASED ON SEQUENCE-DEPENDENT STRUCTURAL FEATURES | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Brown, D.G, Sanderson, M.R, Garman, E, Neidle, S. | | Deposit date: | 1992-03-02 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features.
J.Mol.Biol., 226, 1992
|
|
8A06
 
 | |
8A5T
 
 | |
