1ZQQ
 
 | |
4ZXP
 
 | | Crystal structure of Peptidyl- tRNA Hydrolase from Vibrio cholerae | | Descriptor: | CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Shahid, S, Pal, R.K, Kabra, A, Yadav, R, Kumar, A, Arora, A. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.
RNA, 23, 2017
|
|
3SE0
 
 | | Structural characterization of the subunit A mutant F508W of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2011-06-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Engineered tryptophan in the adenine-binding pocket of catalytic subunit A of A-ATP synthase demonstrates the importance of aromatic residues in adenine binding, forming a tool for steady-state and time-resolved fluorescence spectroscopy.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3SDZ
 
 | | Structural characterization of the subunit A mutant F427W of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2011-06-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Engineered tryptophan in the adenine-binding pocket of catalytic subunit A of A-ATP synthase demonstrates the importance of aromatic residues in adenine binding, forming a tool for steady-state and time-resolved fluorescence spectroscopy.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2NAF
 
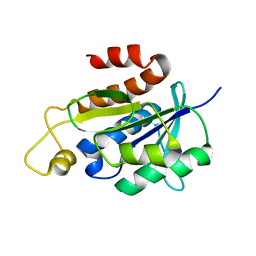 | | Solution structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Fatma, F, Kabra, A, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2015-12-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of peptidyl-tRNA hydrolase from Mycobacterium smegmatis by NMR spectroscopy.
Biochim.Biophys.Acta, 1864, 2016
|
|
6A8M
 
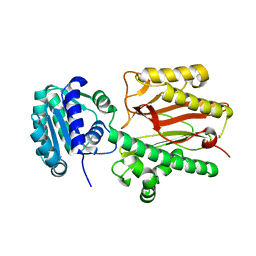 | | N-terminal domain of FACT complex subunit SPT16 from Eremothecium gossypii (Ashbya gossypii) | | Descriptor: | FACT complex subunit SPT16 | | Authors: | Gaur, N.K, Are, V.N, Durani, V, Ghosh, B, Kumar, A, Kulkarni, K, Makde, R.D. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolutionary conservation of protein dynamics: insights from all-atom molecular dynamics simulations of 'peptidase' domain of Spt16.
J.Biomol.Struct.Dyn., 2021
|
|
6B4M
 
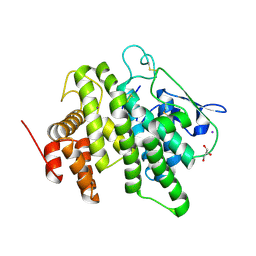 | | Structural characterization of a novel monotreme-specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Sharp, J.A, Kumar, A, Nicholas, K.R, Adams, T.E. | | Deposit date: | 2017-09-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
227D
 
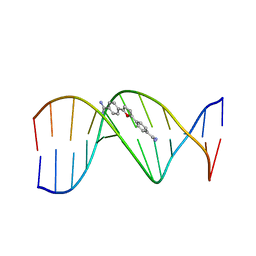 | | A CRYSTALLOGRAPHIC AND SPECTROSCOPIC STUDY OF THE COMPLEX BETWEEN D(CGCGAATTCGCG)2 AND 2,5-BIS(4-GUANYLPHENYL)FURAN, AN ANALOGUE OF BERENIL. STRUCTURAL ORIGINS OF ENHANCED DNA-BINDING AFFINITY | | Descriptor: | 2,5-BIS(4-GUANYLPHENYL)FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Laughton, C.A, Tanious, F, Nunn, C.M, Boykin, D.W, Wilson, W.D, Neidle, S. | | Deposit date: | 1995-08-08 | | Release date: | 1995-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity.
Biochemistry, 35, 1996
|
|
6WY5
 
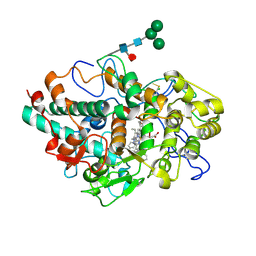 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-37 A.K.A 7-(1-phenyl-3-(((1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl)amino)propyl)-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1-phenyl-3-{[(1S,3S)-3-phenyl-2,3-dihydro-1H-inden-1-yl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WY0
 
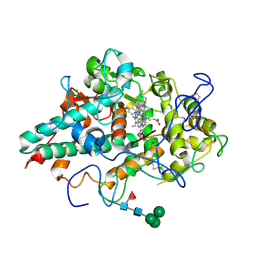 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-40 A.K.A 7-[(1R)-1-phenyl-3-{[(1r,4r)-4-phenylcyclohexyl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(trans-4-phenylcyclohexyl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WXZ
 
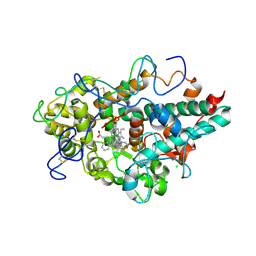 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-29 A.K.A 7-(1,2-DIPHENYLETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1,2-diphenylethyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WYD
 
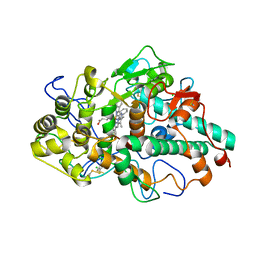 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-12 (AKA; 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyrid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6MVL
 
 | |
6WY7
 
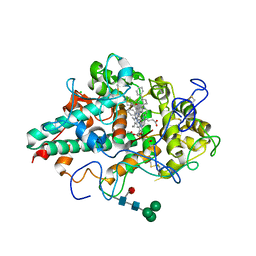 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-41 A.K.A 7-[1-phenyl-3-({4-phenylbicyclo[2.2.2]octan-1-yl}amino)propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(4-phenylbicyclo[2.2.2]octan-1-yl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
1DSJ
 
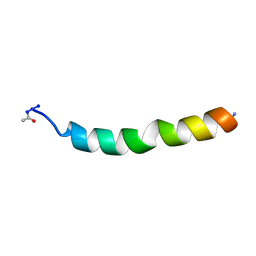 | | NMR SOLUTION STRUCTURE OF VPR50_75, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Torres, A.M, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1997-10-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Helical structure of polypeptides from the C-terminal half of HIV-1 VPR.
Protein Pept.Lett., 5, 1998
|
|
5ITH
 
 | |
