9FTT
 
 | |
1DMG
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L4 | | Descriptor: | CITRIC ACID, RIBOSOMAL PROTEIN L4 | | Authors: | Worbs, M, Huber, R, Wahl, M.C. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon.
EMBO J., 19, 2000
|
|
1FIU
 
 | | TETRAMERIC RESTRICTION ENDONUCLEASE NGOMIV IN COMPLEX WITH CLEAVED DNA | | Descriptor: | ACETIC ACID, DNA (5'-D(*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*CP*GP*C)-3'), ... | | Authors: | Deibert, M, Grazulis, S, Sasnauskas, G, Siksnys, V, Huber, R. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Nat.Struct.Biol., 7, 2000
|
|
1FS8
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-SULFATE COMPLEX | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FS7
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FS9
 
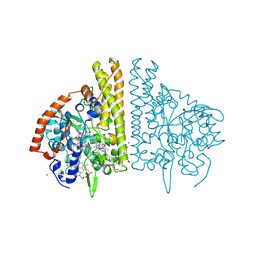 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-AZIDE COMPLEX | | Descriptor: | AZIDE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1DD3
 
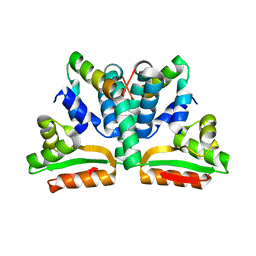 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
1FBX
 
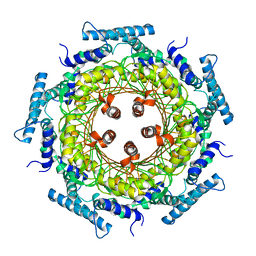 | | CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I | | Descriptor: | CHLORIDE ION, GTP CYCLOHYDROLASE I, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, A, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-17 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FB1
 
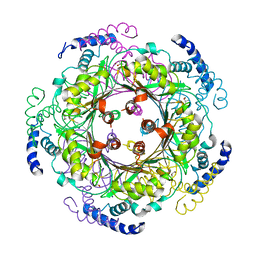 | | CRYSTAL STRUCTURE OF HUMAN GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, G, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ELQ
 
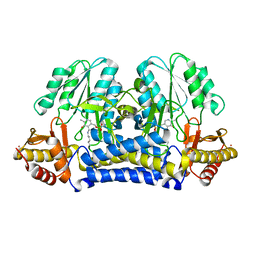 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1G8F
 
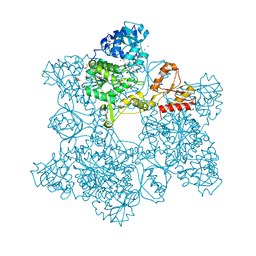 | | ATP SULFURYLASE FROM S. CEREVISIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1DGJ
 
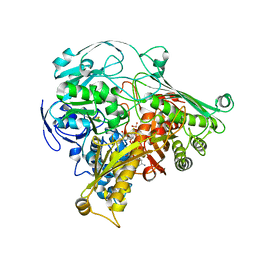 | | CRYSTAL STRUCTURE OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ALDEHYDE OXIDOREDUCTASE, FE2/S2 (INORGANIC) CLUSTER, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Rebelo, J.M, Macieira, S, Dias, J.M, Huber, R, Romao, M.J. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774.
J.Mol.Biol., 297, 2000
|
|
1G63
 
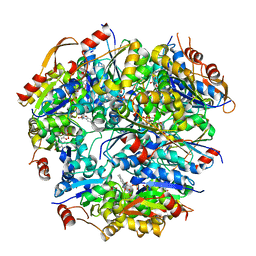 | | PEPTIDYL-CYSTEINE DECARBOXYLASE EPID | | Descriptor: | EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbac, S. | | Deposit date: | 2000-11-03 | | Release date: | 2001-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1G71
 
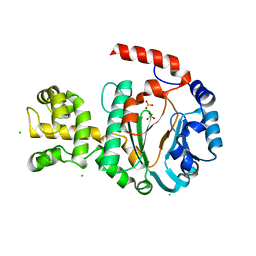 | | CRYSTAL STRUCTURE OF PYROCOCCUS FURIOSUS DNA PRIMASE | | Descriptor: | CHLORIDE ION, DNA PRIMASE, SULFATE ION, ... | | Authors: | Augustin, M.A, Huber, R, Kaiser, J.T. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a DNA-dependent RNA polymerase (DNA primase).
Nat.Struct.Biol., 8, 2001
|
|
1G8G
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE BINARY PRODUCT COMPLEX WITH APS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1EG5
 
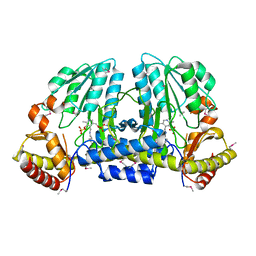 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1ELU
 
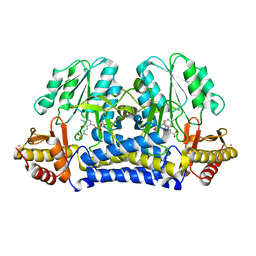 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1G0U
 
 | | A GATED CHANNEL INTO THE PROTEASOME CORE PARTICLE | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Bajorek, M, Kohler, A, Moroder, L, Rubin, D.M, Huber, R, Glickman, M.H, Finley, D. | | Deposit date: | 2000-10-09 | | Release date: | 2000-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gated channel into the proteasome core particle.
Nat.Struct.Biol., 7, 2000
|
|
1G8H
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE TERNARY PRODUCT COMPLEX WITH APS AND PPI | | Descriptor: | ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1CJA
 
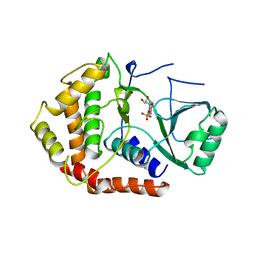 | | ACTIN-FRAGMIN KINASE, CATALYTIC DOMAIN FROM PHYSARUM POLYCEPHALUM | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN (ACTIN-FRAGMIN KINASE) | | Authors: | Steinbacher, S, Hof, P, Eichinger, L, Schleicher, M, Gettemans, J, Vandekerckhove, J, Huber, R, Benz, J. | | Deposit date: | 1999-04-08 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Physarum polycephalum actin-fragmin kinase: an atypical protein kinase with a specialized substrate-binding domain.
EMBO J., 18, 1999
|
|
1CZT
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1CZS
 
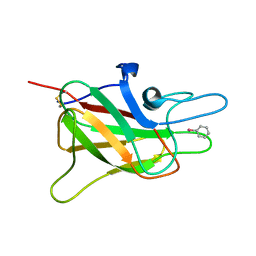 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: COMPLEX WITH PHENYLMERCURY | | Descriptor: | PHENYLMERCURY, PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1EJN
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN INHIBITOR COMPLEX | | Descriptor: | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Sperl, S, Jacob, U, Arroyo de Prada, N, Stuerzebecher, J, Wilhelm, O.G, Bode, W, Magdolen, V, Huber, R, Moroder, L. | | Deposit date: | 2000-04-22 | | Release date: | 2000-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (4-aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E0F
 
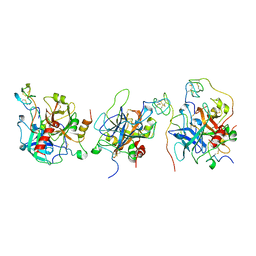 | | Crystal structure of the human alpha-thrombin-haemadin complex: an exosite II-binding inhibitor | | Descriptor: | HAEMADIN, THROMBIN | | Authors: | Richardson, J.L, Kroeger, B, Hoefken, W, Pereira, P, Huber, R, Bode, W, Fuentes-Prior, P. | | Deposit date: | 2000-03-27 | | Release date: | 2000-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Human Alpha-Thrombin-Haemadin Complex: An Exosite II-Binding Inhibitor
Embo J., 19, 2000
|
|
1DXG
 
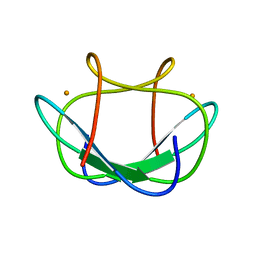 | |
