4N88
 
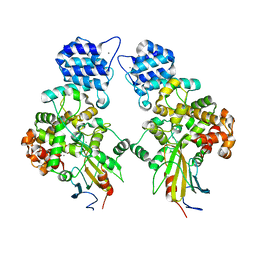 | | Crystal structure of Tse3-Tsi3 complex with calcium ion | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
6IGZ
 
 | | Structure of PSI-LHCI | | Descriptor: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Xiong, P, Xiaochun, Q. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
4N80
 
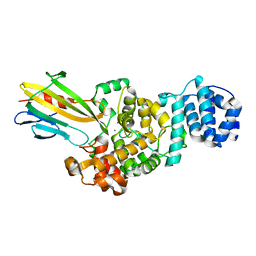 | | Crystal structure of Tse3-Tsi3 complex | | Descriptor: | CALCIUM ION, Uncharacterized protein, ZINC ION | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
4N7S
 
 | | Crystal structure of Tse3-Tsi3 complex with Zinc ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
3I81
 
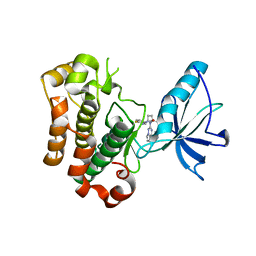 | | Crystal structure of insulin-like growth factor 1 receptor (IGF-1R-WT) complex with BMS-754807 [1-(4-((5-cyclopropyl-1H-pyrazol-3-yl)amino)pyrrolo[2,1-f][1,2,4]triazin-2-yl)-N-(6-fluoro-3-pyridinyl)-2-methyl-L-prolinamide] | | Descriptor: | 1-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}-N-(6-fluoropyridin-3-yl)-2-methyl-L-proli namide, Insulin-like growth factor 1 receptor | | Authors: | Sack, J.S. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development.
J.Med.Chem., 52, 2009
|
|
5ZZD
 
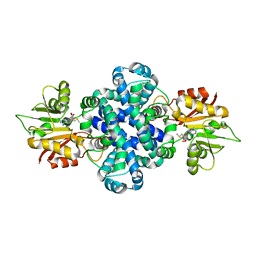 | | Crystal structure of a protein from Aspergillus flavus | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Chang, Z.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-31 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of LepI, a multifunctional SAM-dependent enzyme which catalyzes pericyclic reactions in leporin biosynthesis.
Org.Biomol.Chem., 17, 2019
|
|
7BY7
 
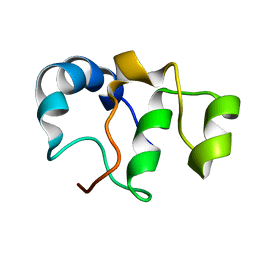 | | Bacteriophage SPO1 protein Gp46 | | Descriptor: | Putative gene 46 protein | | Authors: | Liu, B, Zhang, P. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5V80
 
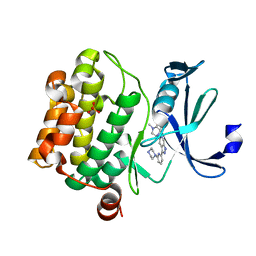 | |
5V82
 
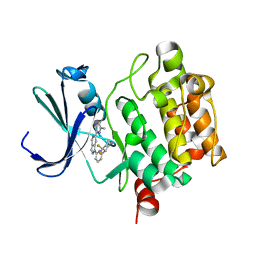 | | PIM1 kinase in complex with Cpd17 (1-(6-(4,4-difluoropiperidin-3-yl)pyridin-2-yl)-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine) | | Descriptor: | 1-{6-[(3R)-4,4-difluoropiperidin-3-yl]pyridin-2-yl}-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Discovery of 5-Azaindazole (GNE-955) as a Potent Pan-Pim Inhibitor with Optimized Bioavailability.
J. Med. Chem., 60, 2017
|
|
4M5E
 
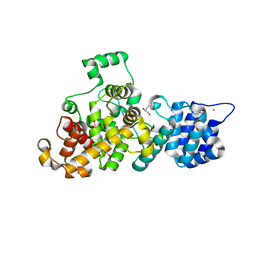 | | Tse3 structure | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Qian, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4M5F
 
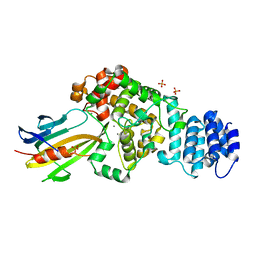 | | complex structure of Tse3-Tsi3 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | Authors: | Gu, L.C. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
2LUQ
 
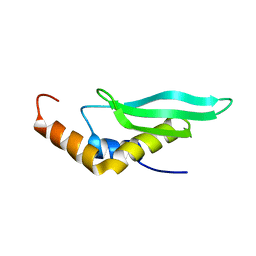 | |
8JUL
 
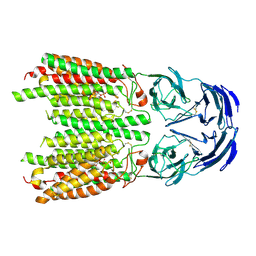 | | Cryo-EM structure of SIDT1 in complex with phosphatidic acid | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
8JUN
 
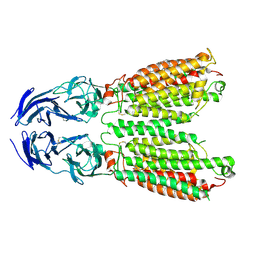 | | Cryo-EM structure of SIDT1 E555Q mutant | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
8GPY
 
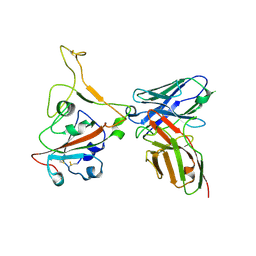 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8IK0
 
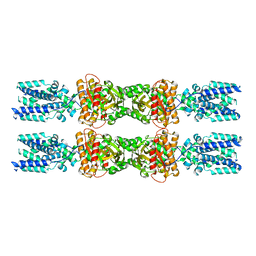 | |
8IK3
 
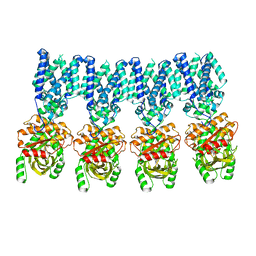 | |
4ZIM
 
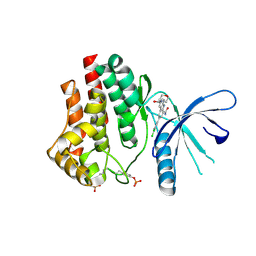 | |
3FM9
 
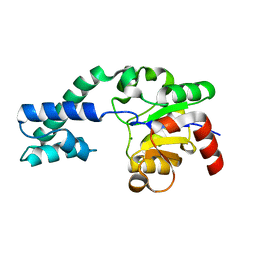 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
7E0B
 
 | |
6LPA
 
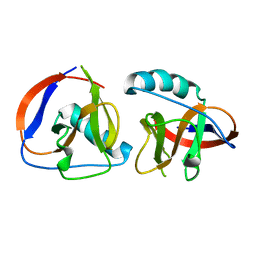 | | The nsp1 protein of a new porcine coronavirus | | Descriptor: | sp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural and Biological Basis of Alphacoronavirus nsp1 Associated with Host Proliferation and Immune Evasion.
Viruses, 12, 2020
|
|
7YJ3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
