7YXR
 
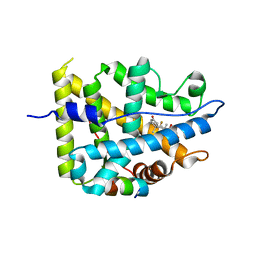 | | Crystal structure of mutant AncGR2-LBD (Y545A) bound to dexamethasone and SHP coregulator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, FORMIC ACID, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXO
 
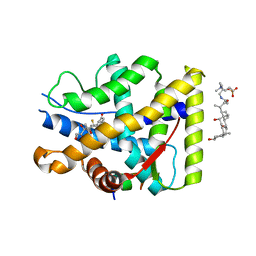 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXN
 
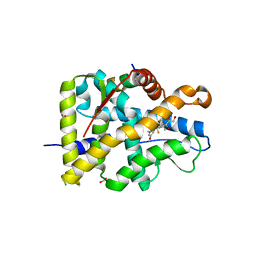 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, FORMIC ACID, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
4P59
 
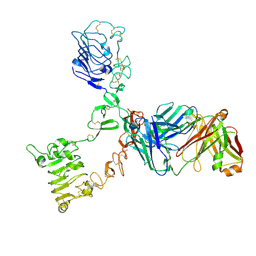 | | HER3 extracellular domain in complex with Fab fragment of MOR09825 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOR09825 Fab fragment heavy chain, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin.
Cancer Res., 73, 2013
|
|
3G0A
 
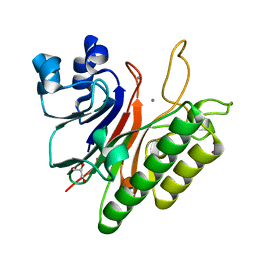 | | Mth0212 with two bound manganese ions | | Descriptor: | Exodeoxyribonuclease, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G2C
 
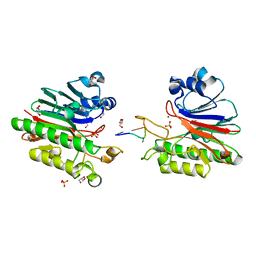 | | Mth0212 in complex with a short ssDNA (CGTA) | | Descriptor: | 5'-D(P*CP*GP*TP*A)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G4T
 
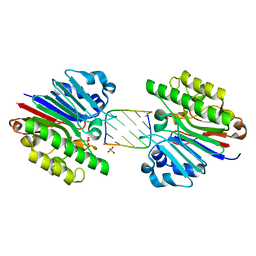 | | Mth0212 (WT) in complex with a 7bp dsDNA | | Descriptor: | 5'-D(*CP*G*TP*AP*CP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-04 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G3C
 
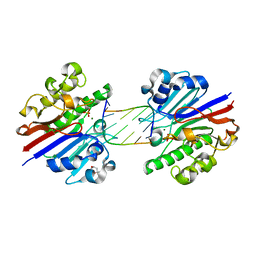 | | Mth0212 (WT) in complex with a 6bp dsDNA containing a single one nucleotide long 3'-overhang | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3FZI
 
 | |
7V4M
 
 | |
7V4O
 
 | | Unique non-heme hydroxylase in biosynthesis of nucleoside antibiotic pathway uncover mechanism of reaction | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, Beta-hydroxylase | | Authors: | Li, T.L, Saeid, M.Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | beta-Hydroxylation of alpha-amino-beta-hydroxylbutanoyl-glycyluridine catalyzed by a nonheme hydroxylase ensures the maturation of caprazamycin
Commun Chem, 5, 2022
|
|
7V4N
 
 | |
7V4P
 
 | |
7V4F
 
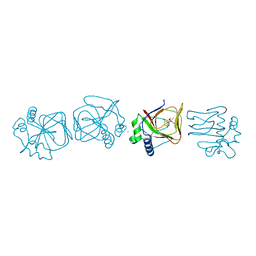 | |
3GA6
 
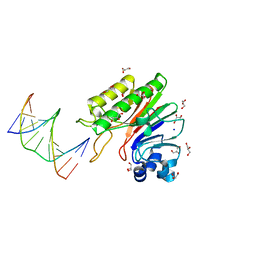 | | Mth0212 in complex with two DNA helices | | Descriptor: | 5'-D(*GP*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
7XE7
 
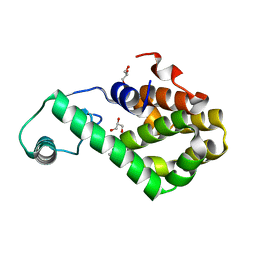 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH10 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE9
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 20% | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Endolysin, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XEA
 
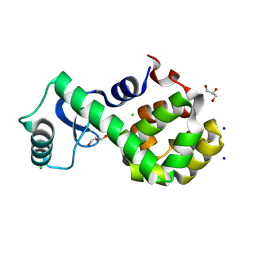 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 40%, and then backsoaking | | Descriptor: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
3G3Y
 
 | | Mth0212 in complex with ssDNA in space group P32 | | Descriptor: | 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-03 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
3G38
 
 | | The catalytically inactive mutant Mth0212 (D151N) in complex with an 8 bp dsDNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*GP*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G1K
 
 | |
3G0R
 
 | | Complex of Mth0212 and an 8bp dsDNA with distorted ends | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
7XE6
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH7 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE5
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH4 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
