7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ7
 
 | | Template lasso peptide C24 mutant F15Y | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
6SM8
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-6-[(3~{S})-3-[(1~{S})-2-cyano-1-[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]ethyl]pyrrolidin-1-yl]benzenecarbonitrile, Tyrosine-protein kinase JAK1 | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6SMB
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | Tyrosine-protein kinase JAK1, ~{N}-[3-[2-[(3-methoxy-1-methyl-pyrazol-4-yl)amino]-5-methyl-pyrimidin-4-yl]-1~{H}-indol-7-yl]-2-methyl-pyridine-3-carboxamide | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
5GMZ
 
 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
7CU6
 
 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
8OWG
 
 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OVZ
 
 | | Crystal structure of D1228V c-MET bound by compound 16 | | Descriptor: | 1-[(1S)-1-[3-(1H-imidazol-4-yl)phenyl]ethyl]-5-(1H-indazol-7-yl)pyrimidine-2,4-dione, Hepatocyte growth factor receptor, IODIDE ION | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUV
 
 | | Crystal structure of D1228V c-MET bound by compound 15 | | Descriptor: | 5-(1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUU
 
 | | Crystal structure of D1228V c-MET bound by compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-ethynyl-5-fluoranyl-1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, FORMIC ACID, ... | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OW3
 
 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OV7
 
 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8Q6M
 
 | | Human SOD1 low dose data collecton | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-08-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
5QTT
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-{[5-amino-1-(3-chloro-2-fluorophenyl)-1H-pyrazole-4-carbonyl]amino}-3-methyl-2-oxo-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Potent, Orally Bioavailable and Efficacious Macrocyclic Inhibitors of Factor XIa. Discovery of Pyridine-Based Macrocycles Possessing Phenylazole Carboxamide P1 Groups.
J.Med.Chem., 63, 2019
|
|
5QTU
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-{[1-(3-chloro-2-fluorophenyl)-5-methyl-1H-imidazole-4-carbonyl]amino}-3-methyl-2-oxo-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Potent, Orally Bioavailable and Efficacious Macrocyclic Inhibitors of Factor XIa. Discovery of Pyridine-Based Macrocycles Possessing Phenylazole Carboxamide P1 Groups.
J.Med.Chem., 63, 2019
|
|
6Y1Y
 
 | |
8X82
 
 | |
8X83
 
 | |
8X84
 
 | |
6KUR
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
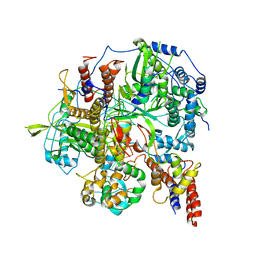 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
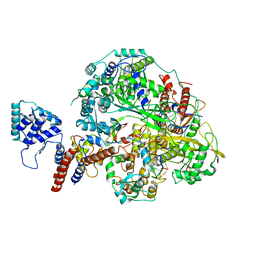 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | Descriptor: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
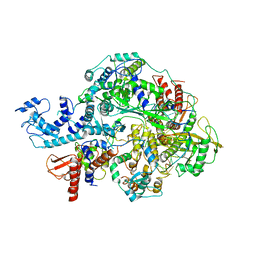 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
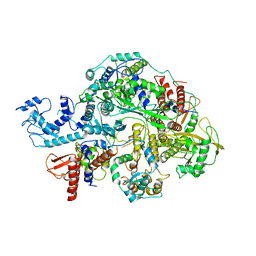 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
