7E4D
 
 | | Crystal structure of PlDBR | | Descriptor: | Double Bond Reductase | | Authors: | Sugimoto, K, Senda, M, Senda, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
5TNH
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 17,18-EpETE hydrolysis intermediate | | Descriptor: | (5Z,8Z,11Z,14Z,17R,18R)-17,18-dihydroxyicosa-5,8,11,14-tetraenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNS
 
 | |
5TNF
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 19,20-EpDPE hydrolysis intermediate | | Descriptor: | (4Z,7Z,10Z,13Z,16Z,19R,20R)-19,20-dihydroxydocosa-4,7,10,13,16-pentaenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNQ
 
 | |
5TNN
 
 | |
5TNJ
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 4-Vinyl-1-cyclohexene 1,2-epoxide hydrolysis intermediate | | Descriptor: | (1R,2R,4R)-4-ethenylcyclohexane-1,2-diol, (1R,2R,4S)-4-ethenylcyclohexane-1,2-diol, (1S,2S,4R)-4-ethenylcyclohexane-1,2-diol, ... | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNK
 
 | |
5TNL
 
 | |
5TND
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 1,2-Epoxycyclohexane hydrolysis intermediate | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, (1S,2S)-cyclohexane-1,2-diol, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNM
 
 | |
5TNE
 
 | |
5TNI
 
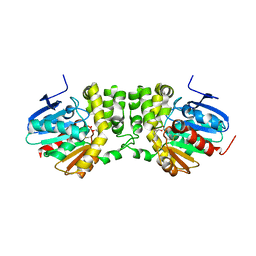 | |
8E1C
 
 | |
8E1B
 
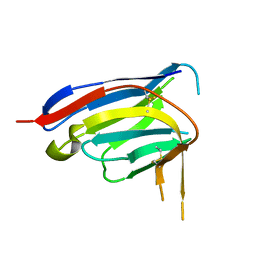 | |
8E2N
 
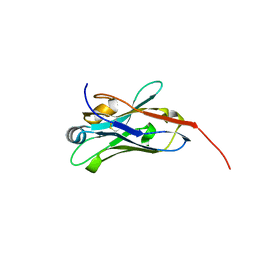 | |
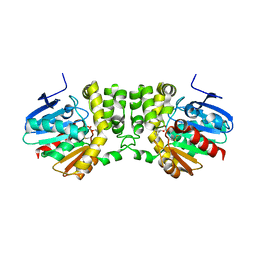
5TNP
 
 | |
5TNG
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EpETE hydrolysis intermediate | | Descriptor: | (5Z,8Z,11Z,14R,15R,17Z)-14,15-dihydroxyicosa-5,8,11,17-tetraenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNR
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 16,17-EpDPE hydrolysis intermediate | | Descriptor: | (4Z,7Z,10Z,13Z,16R,17R,19Z)-16,17-dihydroxydocosa-4,7,10,13,19-pentaenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
8EE2
 
 | |
8EVD
 
 | |
4IW3
 
 | | Crystal structure of a Pseudomonas putida prolyl-4-hydroxylase (P4H) in complex with elongation factor Tu (EF-Tu) | | Descriptor: | Elongation factor Tu-A, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IU2
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cell-wall anchoring protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4IU3
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Cellulose-binding protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
8F6U
 
 | |
