7C8T
 
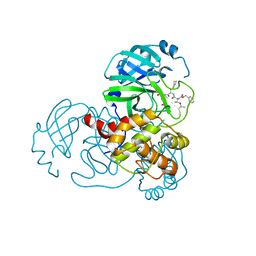 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0205221 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
7CGA
 
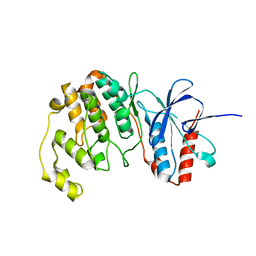 | |
7C8R
 
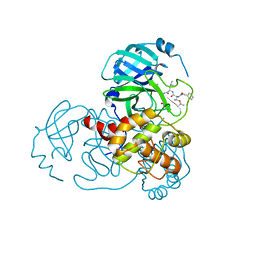 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0203770 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
4ONC
 
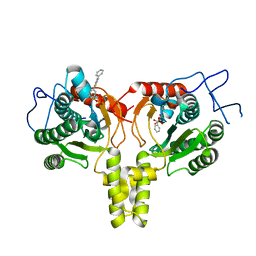 | | Crystal Structure of Mycobacterium Tuberculosis Decaprenyl Diphosphate Synthase in Complex with BPH-640 | | Descriptor: | Decaprenyl diphosphate synthase, [hydroxy(1,1':3',1''-terphenyl-3-yl)methanediyl]bis(phosphonic acid) | | Authors: | Feng, X, Chan, H.C, Ko, T.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Inhibition of Tuberculosinol Synthase and Decaprenyl Diphosphate Synthase from Mycobacterium tuberculosis
J.Am.Chem.Soc., 136, 2014
|
|
1VQA
 
 | |
1VQH
 
 | |
1VQG
 
 | |
1VQD
 
 | |
1VQE
 
 | |
1VQC
 
 | |
1RK6
 
 | | The enzyme in complex with 50mM CdCl2 | | Descriptor: | ACETATE ION, CADMIUM ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1RJR
 
 | | The crystal structure of the D-aminoacylase D366A mutant in complex with 100mM ZnCl2 | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1RJQ
 
 | | The crystal structure of the D-aminoacylase mutant D366A | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1RJP
 
 | | Crystal structure of D-aminoacylase in complex with 100mM CuCl2 | | Descriptor: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1RK5
 
 | | The D-aminoacylase mutant D366A in complex with 100mM CuCl2 | | Descriptor: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1V51
 
 | | The functional role of the binuclear metal center in D-aminoacylase. One-metal activation and second-metal attenuation | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1V4Y
 
 | | The functional role of the binuclear metal center in D-aminoacylase. One-metal activation and second-metal attenuation | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Functional Role of the Binuclear Metal Center in D-Aminoacylase: ONE-METAL ACTIVATION AND SECOND-METAL ATTENUATION.
J.Biol.Chem., 279, 2004
|
|
5B0J
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-undecyl maltoside | | Descriptor: | MoeN5,DNA-binding protein 7d, UNDECYL-MALTOSIDE | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0M
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-dodecyl maltoside | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0K
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-decyl maltoside | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0I
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-octyl glucoside | | Descriptor: | MoeN5,DNA-binding protein 7d, octyl beta-D-glucopyranoside | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B02
 
 | | Structure of the prenyltransferase MoeN5 with a fusion protein tag of Sso7d | | Descriptor: | MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B03
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B00
 
 | | Structure of the prenyltransferase MoeN5 in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, MoeN5 | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0L
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-nonyl glucoside | | Descriptor: | MoeN5,DNA-binding protein 7d, nonyl beta-D-glucopyranoside | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
