3VSY
 
 | |
5NJO
 
 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6Z4A
 
 | |
3VGN
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 3-fluoro-4-nitrophenol | | Descriptor: | 3-fluoro-4-nitrophenol, Steroid Delta-isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P.A. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8UX7
 
 | | Dioclea megacarpa lectin (DmegA) complexed with X-Man | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea megacarpa lectin, ... | | Authors: | Oliveira, M.V, De Sloover, G, Osterne, V.J.S, Pinto-Junior, V.R, Sacramento-Neto, J.C, Van Damme, E.J.M, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2023-11-09 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dioclea megacarpa lectin (DmegA) complexed with X-Man
To Be Published
|
|
8UF4
 
 | | Crystal structure of wildtype dystroglycan proteolytic domain (juxtamembrane domain) | | Descriptor: | Beta-dystroglycan, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, M.J.M, Shi, K, Hayward, A.N, Uhlens, C, Evans III, R.L, Grant, E, Greenberg, L, Aihara, H, Gordon, W.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Analysis of Dystroglycan Cell Surface Cleavage Reveals a Novel Regulation Mechanism
To Be Published
|
|
6QGM
 
 | | VirX1 apo structure | | Descriptor: | VirX1 | | Authors: | Gkotsi, D.S, Ludewig, H, Sharma, S.V, Unsworth, W.P, Taylor, R.J.K, McLachlan, M.M.W, Shanahan, S, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A marine viral halogenase that iodinates diverse substrates.
Nat.Chem., 11, 2019
|
|
8XF7
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XEU
 
 | |
8XFA
 
 | | Structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, FE (III) ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XFI
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound (crystal form 2) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, CHLORIDE ION, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XET
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF4
 
 | | Structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with Bisucaberin bound | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Iron-hydroxamate ABC transporter substrate-binding protein FtsB | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF8
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine B | | Descriptor: | 1,2-ETHANEDIOL, Ferrioxamine B, Iron-hydroxamate ABC transporter substrate-binding protein FtsB, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF9
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
7JW9
 
 | | Ternary cocrystal structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, SODIUM ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K64
 
 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JYB
 
 | | Binary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JV3
 
 | |
6F4C
 
 | | Nicotiana benthamiana alpha-galactosidase | | Descriptor: | alpha-galactosidase | | Authors: | Kytidou, K, Aerts, J.M.F.G, Pannu, N.S. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nicotiana benthamianaalpha-galactosidase A1.1 can functionally complement human alpha-galactosidase A deficiency associated with Fabry disease.
J. Biol. Chem., 293, 2018
|
|
5A8F
 
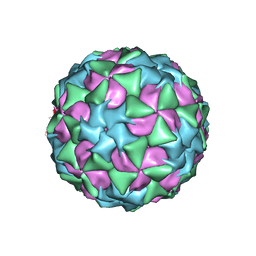 | | Structure and genome release mechanism of human cardiovirus Saffold virus-3 | | Descriptor: | GENOME POLYPHUMAN SAFFOLD VIRUS-3 VP3 PROTEIN, HUMAN SAFFOLD VIRUS-3 VP1, HUMAN SAFFOLD VIRUS-3 VP2 | | Authors: | Mullapudi, E, Novacek, J, Palkova, L, Kulich, P, Lindberg, M, vanKuppeveld, F.J.M, Plevka, P. | | Deposit date: | 2015-07-15 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and Genome Release Mechanism of Human Cardiovirus Saffold Virus-3.
J.Virol., 90, 2016
|
|
6JB2
 
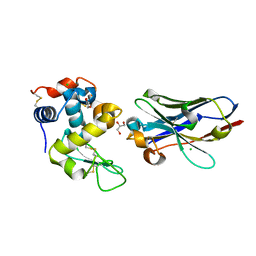 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB9
 
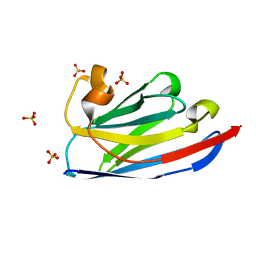 | | Crystal structure of nanobody D3-L11 (unbound form) | | Descriptor: | Nanobody D3-L11, SULFATE ION | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
