1B90
 
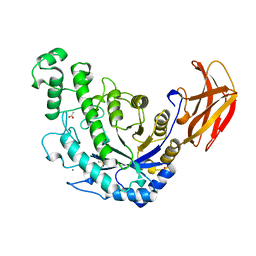 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
6IDR
 
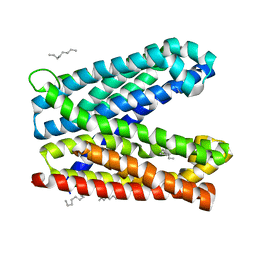 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IDP
 
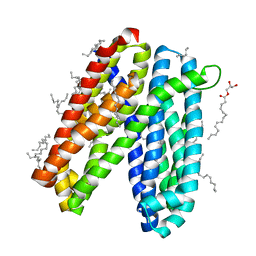 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the straight form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
5ZSU
 
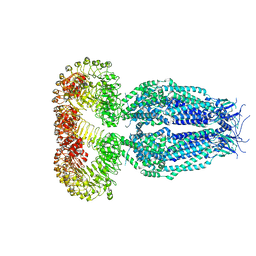 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
1RW4
 
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
1TJ9
 
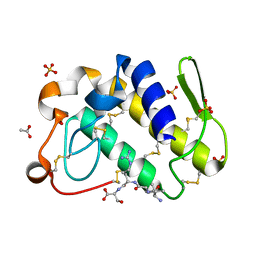 | | Structure of the complexed formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution | | Descriptor: | ACETIC ACID, Phospholipase A2, SULFATE ION, ... | | Authors: | Singh, N, Ethayathulla, A.S, K Somvanshi, R, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Kaur, P, Singh, T.P. | | Deposit date: | 2004-06-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the complex formed between group II phospholipase A2 and a rationally designed tetra peptide,Val-Ala-Arg-Ser at 1.1A resolution
TO BE PUBLISHED
|
|
1TBA
 
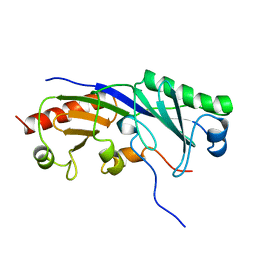 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | Authors: | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
5YIL
 
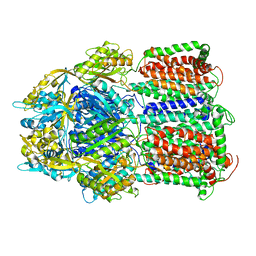 | | Hoisting-loop in bacterial multidrug exporter AcrB is a highly flexible hinge that enables the large motion of the subdomains | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | Zwama, M, Sakurai, K, Hayashi, K, Nakashima, R, Kitagawa, K, Nishino, K, Yamaguchi, A. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hoisting-Loop in Bacterial Multidrug Exporter AcrB Is a Highly Flexible Hinge That Enables the Large Motion of the Subdomains.
Front Microbiol, 8, 2017
|
|
1IRX
 
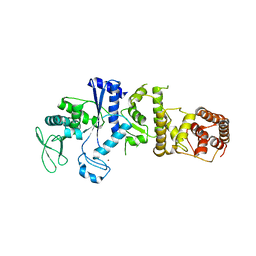 | | Crystal structure of class I lysyl-tRNA synthetase | | Descriptor: | ZINC ION, lysyl-tRNA synthetase | | Authors: | Nureki, O, Terada, T, Ishitani, R, Ambrogelly, A, Ibba, M, Soll, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-10-25 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
Nat.Struct.Biol., 9, 2002
|
|
1IZR
 
 | | F46A mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IVI
 
 | | Crystal Structure of pig dihydrolipoamide dehydrogenase | | Descriptor: | dihydrolipoamide dehydrogenase | | Authors: | Toyoda, T, Kobayashi, R, Sekiguchi, T, Koike, K, Koike, M, Takenaka, A. | | Deposit date: | 2002-03-15 | | Release date: | 2003-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Crystallization and preliminary X-ray analysis of pig E3, lipoamide dehydrogenase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1IZP
 
 | | F46L mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IZQ
 
 | | F46V mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1JSA
 
 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | Descriptor: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | Authors: | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
5XQV
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20L mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XR0
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20T mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2LHI
 
 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
5XQP
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQU
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20I mutant (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
5XUU
 
 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
7JW2
 
 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6AEK
 
 | | Crystal structure of ENPP1 in complex with pApG | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
2MX2
 
 | | UBX-L domain of VCIP135 | | Descriptor: | Deubiquitinating protein VCIP135 | | Authors: | Iwazu, T, Murayama, S, Igarashi, R, Hrioaki, H, Shirakawa, M, Tochio, H. | | Deposit date: | 2014-12-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction mode of the UBX-L domain of VCIP135 determined by solution NMR spectroscopy
To be Published
|
|
