3BJZ
 
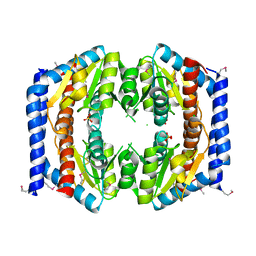 | | Crystal structure of Pseudomonas aeruginosa phosphoheptose isomerase | | Descriptor: | CHLORIDE ION, Phosphoheptose isomerase, SULFATE ION | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants.
J.Biol.Chem., 283, 2008
|
|
5F89
 
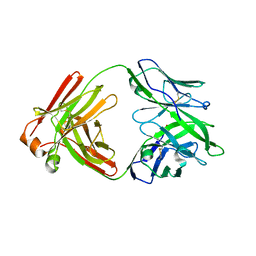 | |
5XP6
 
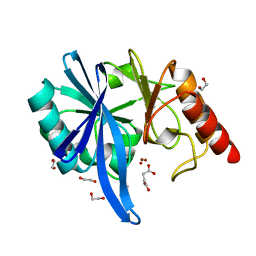 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
5FHY
 
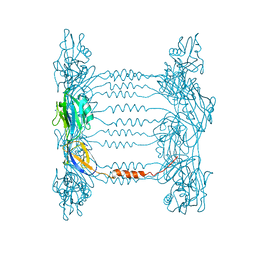 | | Crystal structure of FliD (HAP2) from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellar hook-associated protein 2, SODIUM ION | | Authors: | Postel, S, Bonsor, D, Diederichs, K, Sundberg, E.J. | | Deposit date: | 2015-12-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Bacterial flagellar capping proteins adopt diverse oligomeric states.
Elife, 5, 2016
|
|
5XP9
 
 | | Crystal structure of Bismuth bound NDM-1 | | Descriptor: | Bismuth(III) ION, GLYCEROL, Metallo-beta-lactamase type 2 | | Authors: | Zhang, H, Ma, G, Lai, T.P, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
3CFZ
 
 | |
7SN6
 
 | | U2AF65 UHM BOUND TO SF3B155 ULM5 | | Descriptor: | ISOPROPYL ALCOHOL, Splicing factor 3B subunit 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A UHM-ULM interface with unusual structural features contributes to U2AF2 and SF3B1 association for pre-mRNA splicing.
J.Biol.Chem., 298, 2022
|
|
3KTZ
 
 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3KU0
 
 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3PGK
 
 | | The structure of yeast phosphoglycerate kinase at 0.25 nm resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shaw, P.J, Walker, N.P, Watson, H.C. | | Deposit date: | 1982-07-15 | | Release date: | 1982-09-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence and structure of yeast phosphoglycerate kinase.
Embo J., 1, 1982
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6EEY
 
 | |
6E58
 
 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) | | Descriptor: | CALCIUM ION, Secreted Endo-beta-N-acetylglucosaminidase (EndoS) | | Authors: | Klontz, E.H, Trastoy, B, Gunther, S, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
8FZV
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, expressed with SUMO tag | | Descriptor: | MAGNESIUM ION, Transcription factor ETV6,Anthrax toxin receptor 2, UNKNOWN ATOM OR ION | | Authors: | Pedroza Romo, M.J, Soleimani, S, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3JYT
 
 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with DATP as the incoming nucleotide substrate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
3JSM
 
 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with tenofovir-diphosphate as the incoming nucleotide substrate | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
8YTI
 
 | | Crystal Structure of Nucleosome-H1x Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Qiuye, B, Padavattan, S, Davey, C.A. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Linker Histones Associate Heterogeneously with Nucleosomes in the Condensed State
To Be Published
|
|
1W05
 
 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Alanine-Fe Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-ALANINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
1W06
 
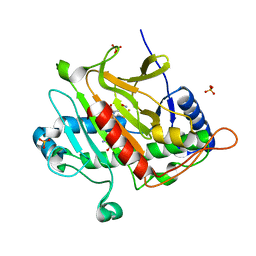 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Alanine-Fe NO Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-ALANINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
1W03
 
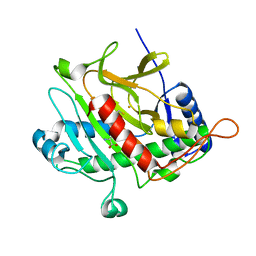 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Glycine-Fe Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-GLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
1W04
 
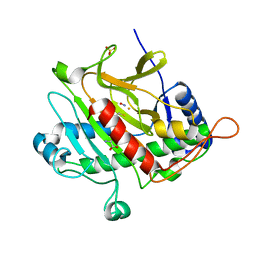 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Glycine-Fe-NO Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-GLYCINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
7Z1P
 
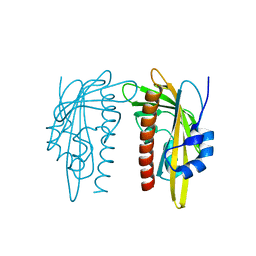 | | X-ray crystal structure of SLPYL1-E151D mutant | | Descriptor: | SLPYL1-E151D | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Modulation of the Ligand Sensitivity of a Tomato Dimeric Abscisic Acid Receptor Through a Glu to Asp Mutation in the Latch Loop.
Front Plant Sci, 13, 2022
|
|
7Z1S
 
 | | X-ray crystal structure of SLPYL1-NIO complex | | Descriptor: | GLYCEROL, NICOTINIC ACID, SlPYL1-NIO | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Modulation of the Ligand Sensitivity of a Tomato Dimeric Abscisic Acid Receptor Through a Glu to Asp Mutation in the Latch Loop.
Front Plant Sci, 13, 2022
|
|
7Z1Q
 
 | |
