7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | Descriptor: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
5YQN
 
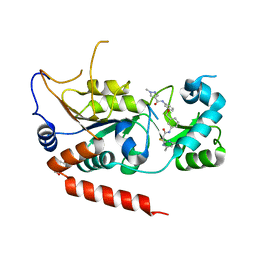 | | Crystal structure of Sirt2 in complex with selective inhibitor L55 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[3-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQO
 
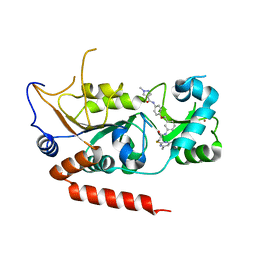 | | Crystal structure of Sirt2 in complex with selective inhibitor L5C | | Descriptor: | N-[4-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQL
 
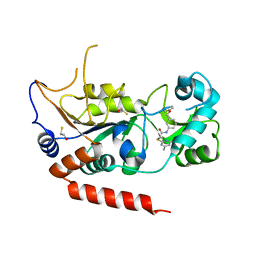 | | Crystal structure of Sirt2 in complex with selective inhibitor A2I | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[3-(phenoxymethyl)phenyl]ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQM
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A29 | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-(4-phenylsulfanylphenyl)ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
4E50
 
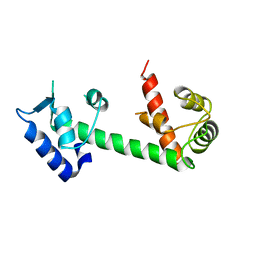 | | Calmodulin and Ng peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neurogranin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
4E53
 
 | | Calmodulin and Nm peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neuromodulin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
1ZDL
 
 | | Crystal Structure of Mouse Thioredoxin Reductase Type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 2, ... | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-04-14 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZKQ
 
 | | Crystal structure of mouse thioredoxin reductase type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 2, mitochondrial | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-05-03 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7VBT
 
 | | Crystal structure of RIOK2 in complex with CQ211 | | Descriptor: | 8-(6-methoxypyridin-3-yl)-1-[4-piperazin-1-yl-3-(trifluoromethyl)phenyl]-5H-[1,2,3]triazolo[4,5-c]quinolin-4-one, Serine/threonine-protein kinase RIO2 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-09-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54001474 Å) | | Cite: | Discovery of 8-(6-Methoxypyridin-3-yl)-1-(4-(piperazin-1-yl)-3-(trifluoromethyl)phenyl)-1,5-dihydro- 4H -[1,2,3]triazolo[4,5- c ]quinolin-4-one (CQ211) as a Highly Potent and Selective RIOK2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7F3N
 
 | | Structure of PopP2 in apo form | | Descriptor: | Type III effector protein popp2 | | Authors: | Xia, Y, Zhang, Z.M. | | Deposit date: | 2021-06-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.351856 Å) | | Cite: | Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase.
Nat Commun, 12, 2021
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
