8K7N
 
 | | Structure of ferric-binding protein KfuA | | Descriptor: | FE (III) ION, Ferric iron ABC transporter, iron-binding protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Klebsiella pneumoniae ferric-binding protein KfuA
To be published
|
|
5Z10
 
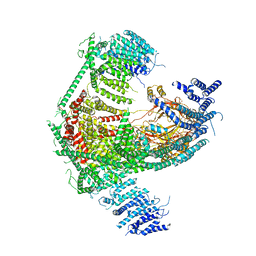 | | Structure of the mechanosensitive Piezo1 channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | Deposit date: | 2017-12-22 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
2A04
 
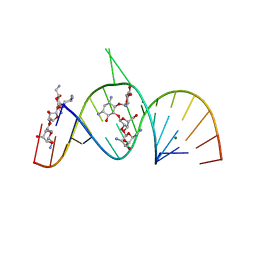 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1ZX7
 
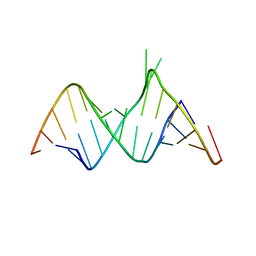 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3' | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1ZZ5
 
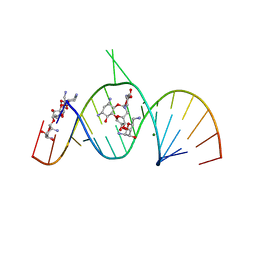 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 13,15-DIAMINO-2-(AMINOMETHYL)-3,4,9,12-TETRAHYDROXYHEXADECAHYDRO-2H-7,10-EPOXYPYRANO[2,3-B][1,10,4]BENZODIOXAZACYCLODODECIN-8-YL 2,6-DIAMINO-2,6-DIDEOXYHEXOPYRANOSIDE, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-13 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
3GYN
 
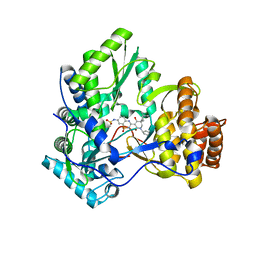 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydropyridinone inhibitor | | Descriptor: | N-{3-[(5R)-1-cyclopentyl-4-hydroxy-5-methyl-5-(3-methylbutyl)-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IGV
 
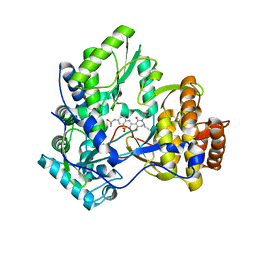 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CDE
 
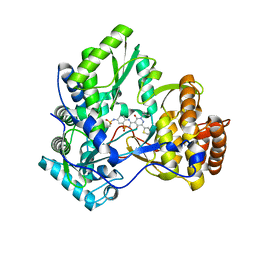 | | Crystal structure of HCV NS5B polymerase with a novel Pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-02-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7'-substituents and initial pharmacokinetic assessments.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D5M
 
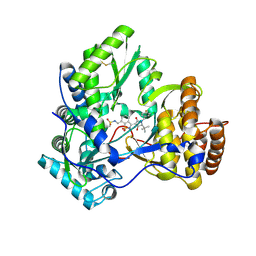 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(3-chloro-4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzis othiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-05-16 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of 1,1-dioxoisothiazole and benzo[b]thiophene-1,1-dioxide derivatives as novel inhibitors of hepatitis C virus NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BR9
 
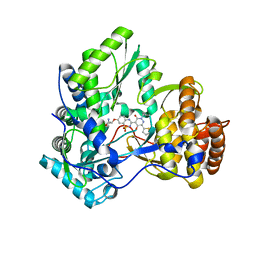 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | (2R)-2-({3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothia diazin-7-yl}oxy)propanamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BSC
 
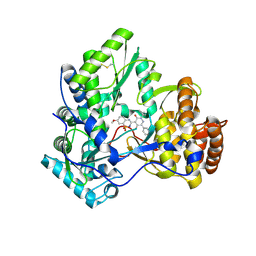 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 5-hydroxy-4-(7-methoxy-1,1-dioxido-2H-1,2,4-benzothiadiazin-3-yl)-2-(3-methylbutyl)-6-phenylpyridazin-3(2H)-one, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CVK
 
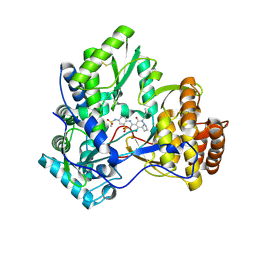 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[1-(3,3-Dimethyl-butyl)-4-hydroxy-2-oxo-1,2,4a,5,6,7-hexahydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,2-dihydro -1lambda6-benzo[1,2,4]thiadiazin-7-yl}-methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-04-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hexahydro-pyrrolo- and hexahydro-1H-pyrido[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5K9I
 
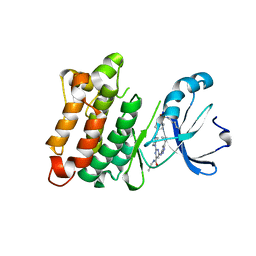 | | Crystal structure of c-SRC in complex with a covalent lysine probe | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Wan, X, Ouyang, S, Zhao, Q, Taunton, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
8W22
 
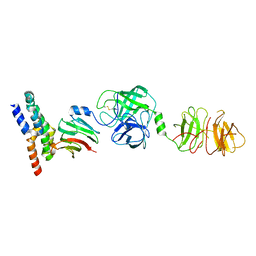 | | Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) | | Descriptor: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | Authors: | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
8W20
 
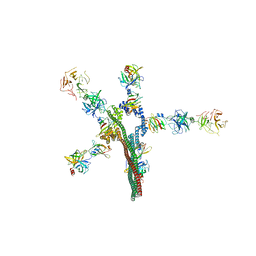 | | Umb1 umbrella toxin particle | | Descriptor: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | Authors: | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
5TGZ
 
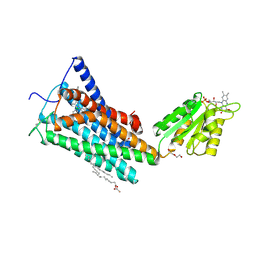 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
2D45
 
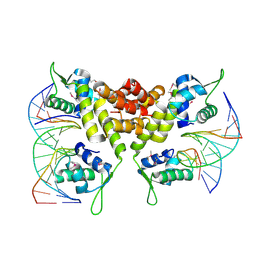 | | Crystal structure of the MecI-mecA repressor-operator complex | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2005-10-09 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the MecI repressor from Staphylococcus aureus in complex with the cognate DNA operator of mec.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3A2A
 
 | | The structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1 | | Descriptor: | CHLORIDE ION, Voltage-gated hydrogen channel 1 | | Authors: | Li, S.J, Unno, H, Zhou, Q, Zhao, Q, Zhai, Y, Sun, F. | | Deposit date: | 2009-05-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1.
J.Biol.Chem., 285, 2010
|
|
1GA5
 
 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*(5IT)P*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
1HLZ
 
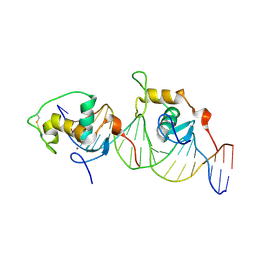 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
4B3N
 
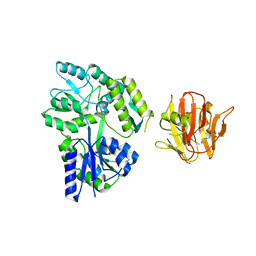 | | Crystal structure of rhesus TRIM5alpha PRY/SPRY domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MALTOSE-BINDING PERIPLASMIC PROTEIN, TRIPARTITE MOTIF-CONTAINING PROTEIN 5, ... | | Authors: | Yang, H, Ji, X, Zhao, Q, Xiong, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insight Into HIV-1 Capsid Recognition by Rhesus Trim5Alpha
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1DSZ
 
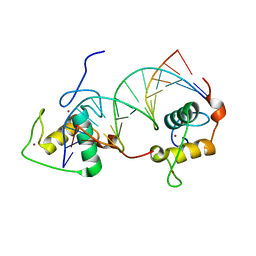 | | STRUCTURE OF THE RXR/RAR DNA-BINDING DOMAIN HETERODIMER IN COMPLEX WITH THE RETINOIC ACID RESPONSE ELEMENT DR1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*G)-3'), RETINOIC ACID RECEPTOR ALPHA, ... | | Authors: | Rastinejad, F, Wagner, T, Zhao, Q, Khorasanizadeh, S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1.
EMBO J., 19, 2000
|
|
6FK5
 
 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to DNA substrate in presence of magnesium ion | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FKE
 
 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to product DNA hairpin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*A)-3'), Exodeoxyribonuclease III, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FK4
 
 | | Structure of 3' phosphatase NExo (WT) from Neisseria bound to DNA substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), Exodeoxyribonuclease III | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
