5GPP
 
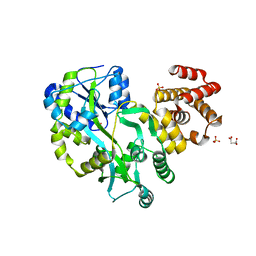 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5GPQ
 
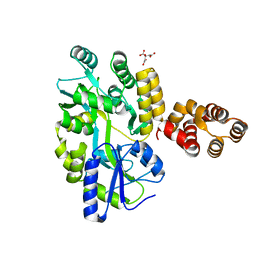 | | Crystal Structure of zebrafish ASC CARD Domain | | Descriptor: | CITRIC ACID, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
7XB4
 
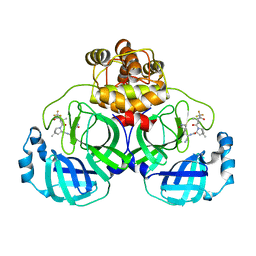 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Hu, X.H, Li, J, Zhang, J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
5GP1
 
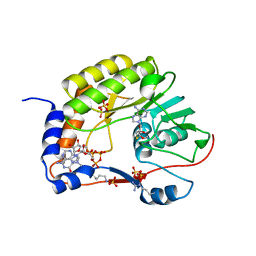 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5GOZ
 
 | |
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
2FVD
 
 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
7XAX
 
 | | Crystal structure of SARS coronavirus main protease in complex with Baicalei | | Descriptor: | 3C-like proteinase nsp5, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-03-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of SARS-CoV 3C-like protease with baicalein.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
4TPY
 
 | | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, ... | | Authors: | Teplitsky, E, Joshi, K, Mullen, J.D, Soares, A.S. | | Deposit date: | 2014-06-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
6KXG
 
 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
6ABS
 
 | | Actin interacting protein 5 (Aip5, mutant) | | Descriptor: | Actin binding protein, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Sun, J, Ying, X, Toh, J, Hong, W, Miao, Y, Gao, Y.G. | | Deposit date: | 2018-07-23 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
1NUE
 
 | | X-RAY STRUCTURE OF NM23 HUMAN NUCLEOSIDE DIPHOSPHATE KINASE B COMPLEXED WITH GDP AT 2 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Morera, S, Lacombe, M.-L, Yingwu, X, Lebras, G, Janin, J. | | Deposit date: | 1995-10-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 A resolution.
Structure, 3, 1995
|
|
