1BZB
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
4H04
 
 | | Lacto-N-biosidase from Bifidobacterium bifidum | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ito, T, Katayama, T, Wada, J, Suzuki, R, Ashida, H, Wakagi, T, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2012-09-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
3JCJ
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
3JCN
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association: Initiation Complex I | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | Descriptor: | CALCITONIN | | Authors: | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
3UES
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UET
 
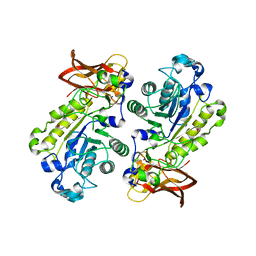 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
6J7I
 
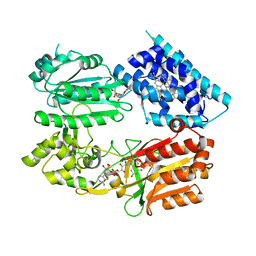 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (15aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
8I04
 
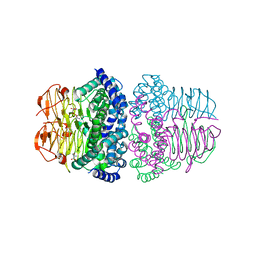 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I09
 
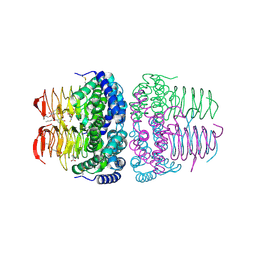 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with butyl gallate | | Descriptor: | CYSTEINE, PHOSPHATE ION, Serine acetyltransferase, ... | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I06
 
 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with CoA | | Descriptor: | COENZYME A, CYSTEINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
6J7A
 
 | | Fusion protein of heme oxygenase-1 and NADPH cytochrome P450 reductase (17aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Sato, H, Wada, K, Yamamoto, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.269 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
2LQI
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LQH
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XUQ
 
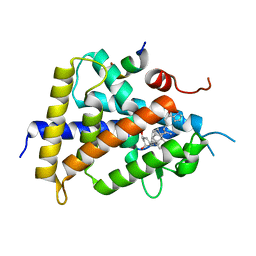 | | Crystal structure of VDR-LBD complexed with an antagonist, 2-methylidene-19,26,27-trinor-22-(S)-butyl-1-hydroxy-25-oxo-25-(1H-pyrrol-2-yl)- vitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(1~{H}-pyrrol-2-yl)octan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent Vitamin D Receptor Antagonist
To Be Published
|
|
5FB3
 
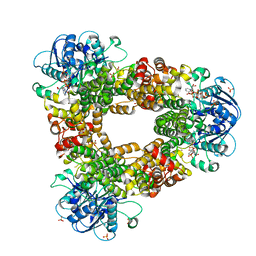 | | Structure of glycerophosphate dehydrogenase in complex with NADPH | | Descriptor: | Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE, ... | | Authors: | Sakuraba, H, Hayashi, J, Yamamoto, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique coenzyme binding mode of hyperthermophilic archaeal sn-glycerol-1-phosphate dehydrogenase from Pyrobaculum calidifontis
Proteins, 84, 2016
|
|
6JF0
 
 | |
6JEZ
 
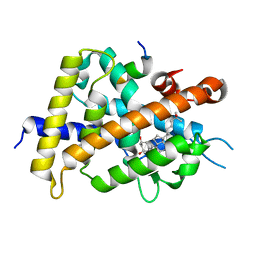 | | Covalent labeling of rVDR-LBD by turn-on fluorescent probe mediated by conjugate addition and cyclization | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, 7-(diethylamino)chromen-2-one, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
2ZXQ
 
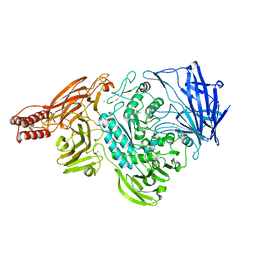 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
6IQ5
 
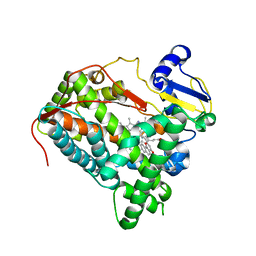 | | Crystal Structure of CYP1B1 and Inhibitor Having Azide Group | | Descriptor: | 2-(cis-4-azidocyclohexyl)-4H-naphtho[1,2-b]pyran-4-one, Cytochrome P450 1B1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kubo, M, Yamamoto, K, Itoh, T. | | Deposit date: | 2018-11-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design and synthesis of selective CYP1B1 inhibitor via dearomatization of alpha-naphthoflavone.
Bioorg. Med. Chem., 27, 2019
|
|
6JEY
 
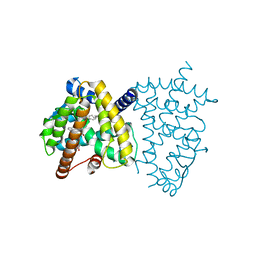 | | Covalent bond formation between ynone moiety of synthetic fatty acid and hPPARg-LBD | | Descriptor: | (9Z,12Z,15Z,18Z,21Z)-5-oxidanylidenetetracosa-9,12,15,18,21-pentaen-6-ynoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
2M33
 
 | | Solution NMR structure of full-length oxidized microsomal rabbit cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Subramanian, V, Ahuja, S, Popovych, N, Huang, R, Le Clair, S.V, Jahr, N, Soong, R, Xu, J, Yamamoto, K, Nanga, R.P, Im, S, Waskell, L, Ramamoorthy, A. | | Deposit date: | 2013-01-08 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of full-length mammalian cytochrome b5
To be Published
|
|
3AHG
 
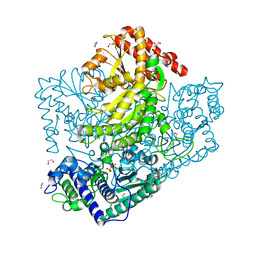 | | H64A mutant of Phosphoketolase from Bifidobacterium Breve complexed with a tricyclic ring form of thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[(9aR)-2,7-dimethyl-9a,10-dihydro-5H-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-8-yl]ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
