8CYZ
 
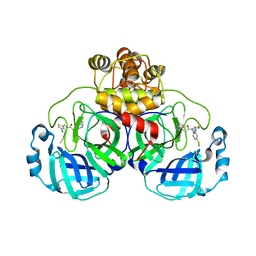 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
4G2S
 
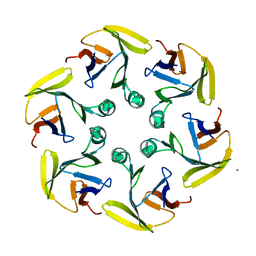 | |
5TZ8
 
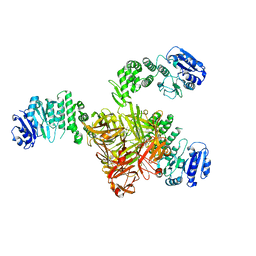 | | Crystal structure of S. aureus TarS | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZK
 
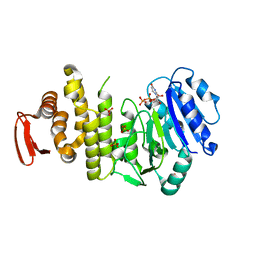 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZI
 
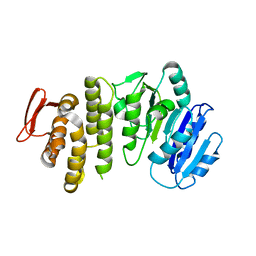 | | Crystal structure of S. aureus TarS 1-349 | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5U02
 
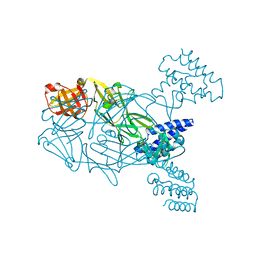 | | Crystal structure of S. aureus TarS 217-571 | | Descriptor: | Glycosyl transferase, IMIDAZOLE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-22 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZJ
 
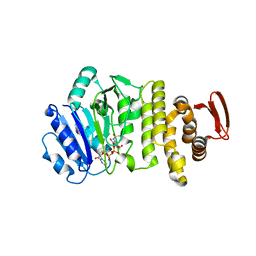 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZE
 
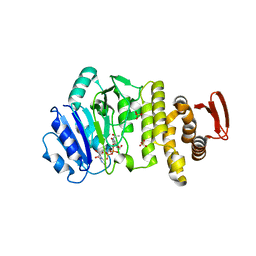 | | Crystal structure of S. aureus TarS in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5WC8
 
 | |
5WC6
 
 | |
3J1W
 
 | |
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NTZ
 
 | | Crystal structure of E. coli PBP5-meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6NTW
 
 | | Crystal structure of E. coli YcbB | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable L,D-transpeptidase YcbB, SULFATE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6NJP
 
 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
4XP8
 
 | |
8EXS
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
4QDD
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with 1,4-30Q-CoA | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QDF
 
 | | Crystal structure of apo KshA5 and KshA1 in complex with 1,4-30Q-CoA from R. rhodochrous | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QDC
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with FE2/S2 (INORGANIC) CLUSTER | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QCK
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis in complex with 4-androstene-3,17-dione | | Descriptor: | 3-ketosteroid-9-alpha-monooxygenase oxygenase subunit, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
6XJI
 
 | | PmtCD ABC exporter at C1 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
