4OTC
 
 | |
5UIF
 
 | | Crystal Structure of Native Ps01740 | | Descriptor: | Ps01740 | | Authors: | LeVieux, J, Baas, B.J, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Kinetic and structural characterization of a cis-3-Chloroacrylic acid dehalogenase homologue in Pseudomonas sp. UW4: A potential step between subgroups in the tautomerase superfamily.
Arch. Biochem. Biophys., 636, 2017
|
|
5UNQ
 
 | |
6BLM
 
 | |
6DAQ
 
 | | PhdJ bound to substrate intermediate | | Descriptor: | PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
6DAN
 
 | | PhdJ WT 2 Angstroms resolution | | Descriptor: | CHLORIDE ION, PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
6DAO
 
 | | NahE WT selenomethionine | | Descriptor: | Trans-O-hydroxybenzylidenepyruvate hydratase-aldolase | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
3MF7
 
 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
3M20
 
 | | Crystal structure of DmpI from Archaeoglobus fulgidus determined to 2.37 Angstroms resolution | | Descriptor: | 4-oxalocrotonate tautomerase, putative | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3M21
 
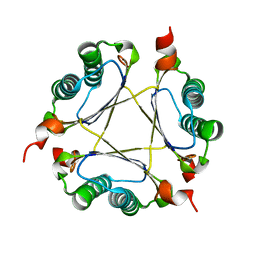 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | Descriptor: | Probable tautomerase HP_0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3N4H
 
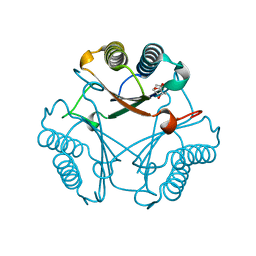 | |
3N4G
 
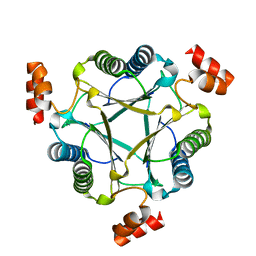 | | Crystal structure of native Cg10062 | | Descriptor: | Putative tautomerase | | Authors: | Guo, Y, Robertson, B.A, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-05-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of the Native and Inactivated Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase from Corynebacterium glutamicum: Implications for the Evolution of cis-3-Chloroacrylic Acid Dehalogenase Activity in the Tautomerase Superfamily
To be Published
|
|
3N4D
 
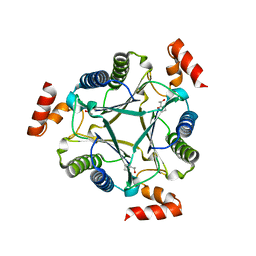 | |
3MLC
 
 | | Crystal structure of FG41MSAD inactivated by 3-chloropropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3MB2
 
 | | Kinetic and Structural Characterization of a Heterohexamer 4-Oxalocrotonate Tautomerase from Chloroflexus aurantiacus J-10-fl: Implications for Functional and Structural Diversity in the Tautomerase Superfamily | | Descriptor: | 4-oxalocrotonate tautomerase family enzyme - alpha subunit, 4-oxalocrotonate tautomerase family enzyme - beta subunit, SULFATE ION | | Authors: | Burks, E.A, Fleming, C.D, Mesecar, A.D, Whitman, C.P, Pegan, S.D. | | Deposit date: | 2010-03-24 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily
Biochemistry, 49, 2010
|
|
3MJZ
 
 | | The crystal structure of native FG41 MSAD | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson, W.H.Jr, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3MF8
 
 | | Crystal Structure of Native cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
8DO5
 
 | | Crystal structure of NahE in complex with intermediate (R)-4-hydroxy-4-(2-hydroxyphenyl)-2-iminobutanoate | | Descriptor: | (4R)-4-hydroxy-4-(2-hydroxyphenyl)butanoic acid, DIMETHYL SULFOXIDE, Trans-ohydrobenzylidenepyruvate hydratase aldolase | | Authors: | LeVieux, J.A, Hardtke, H.A, Zhang, Y.J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mutagenic analysis of NahE, a hydratase-aldolase in the naphthalene degradative pathway.
Arch.Biochem.Biophys., 733, 2023
|
|
7M59
 
 | |
7M58
 
 | |
7TVK
 
 | |
3ITG
 
 | |
2EKG
 
 | |
2FM7
 
 | |
8T9P
 
 | |
