2FUL
 
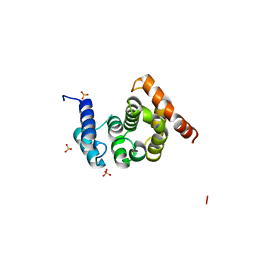 | | Crystal Structure of the C-terminal Domain of S. cerevisiae eIF5 | | Descriptor: | Eukaryotic translation initiation factor 5, SULFATE ION | | Authors: | Wei, Z, Xue, Y, Xu, H, Gong, W. | | Deposit date: | 2006-01-27 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-terminal Domain of S.cerevisiae eIF5
J.Mol.Biol., 359, 2006
|
|
3PZD
 
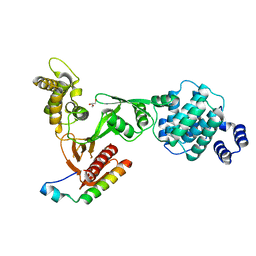 | | Structure of the myosin X MyTH4-FERM/DCC complex | | Descriptor: | GLYCEROL, Myosin-X, Netrin receptor DCC | | Authors: | Wei, Z, Yan, J, Pan, L, Zhang, M. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6IVU
 
 | |
6IVS
 
 | |
9B0Q
 
 | |
9B0G
 
 | |
9B0S
 
 | |
9B0R
 
 | |
9B0H
 
 | |
9B0F
 
 | |
9B0P
 
 | |
4D8O
 
 | |
4G85
 
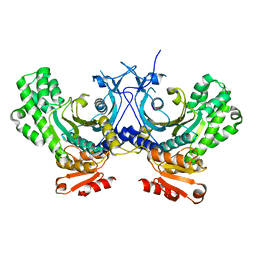 | | Crystal structure of human HisRS | | Descriptor: | Histidine-tRNA ligase, cytoplasmic | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
4G84
 
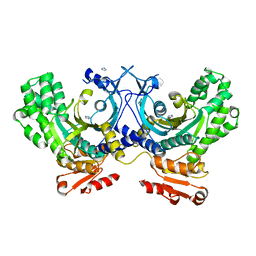 | | Crystal structure of human HisRS | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic, ... | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
4MPL
 
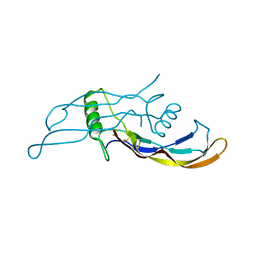 | | Crystal structure of BMP9 at 1.90 Angstrom | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Li, W, Morrell, N.W, Wei, Z. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of Bone Morphogenetic Protein 9 (BMP9) by Redox-dependent Proteolysis.
J.Biol.Chem., 289, 2014
|
|
4P7I
 
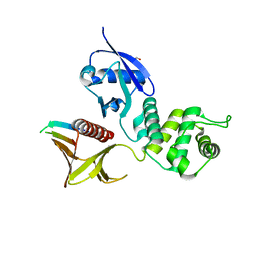 | | Crystal structure of the Merlin FERM/DCAF1 complex | | Descriptor: | GLYCEROL, Merlin, Protein VPRBP | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the binding of Merlin FERM domain to the E3 ubiquitin ligase substrate adaptor DCAF1.
J.Biol.Chem., 289, 2014
|
|
4WSI
 
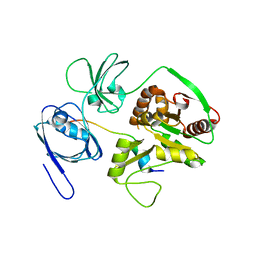 | | Crystal Structure of PALS1/Crb complex | | Descriptor: | MAGUK p55 subfamily member 5, Protein crumbs | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RLV
 
 | |
4RLY
 
 | |
5YAZ
 
 | | Crystal structure of the ANKRD domain of KANK1 | | Descriptor: | ACETATE ION, KN motif and ankyrin repeat domains 1 | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
5YAY
 
 | | Crystal structure of KANK1/KIF21A complex | | Descriptor: | KN motif and ankyrin repeat domains 1, Kinesin-like protein KIF21A | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
3R0H
 
 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
1TXC
 
 | |
4U7U
 
 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
1TW0
 
 | |
