7Y6T
 
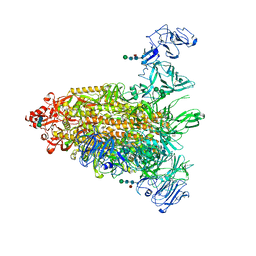 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
6KL5
 
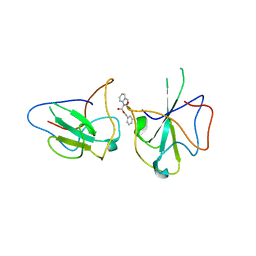 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL6
 
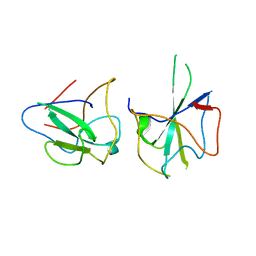 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
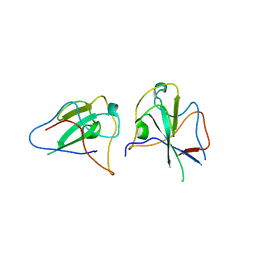 | |
7WCC
 
 | | Oxidase ChaP-D49L/Q91C mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Sun, M.X, Zheng, W, Wang, Y.S, Zhu, J.P, Tan, R.X. | | Deposit date: | 2021-12-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Oxidase ChaP-D49L/Q91C mutant
To Be Published
|
|
3KMY
 
 | | Structure of BACE bound to SCH12472 | | Descriptor: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KMX
 
 | | Structure of BACE bound to SCH346572 | | Descriptor: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | Descriptor: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
7WB2
 
 | | Oxidase ChaP-D49L/Y109F mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Zong, Y, Zheng, W, Wang, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7W5E
 
 | | Oxidase ChaP D49L mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Wang, Y, Zheng, W, Meng, Z, Jin, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6A52
 
 | | Oxidase ChaP-H1 | | Descriptor: | FE (II) ION, dioxidase ChaP-H1 | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | Descriptor: | ChaP protein, FE (II) ION | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
3L59
 
 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L58
 
 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
1FHS
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY DOMAIN-2 OF THE GROWTH FACTOR RECEPTOR BOUND PROTEIN-2, NMR, 18 STRUCTURES | | Descriptor: | GROWTH FACTOR RECEPTOR BOUND PROTEIN-2 | | Authors: | Senior, M.M, Frederick, A.F, Black, S, Perkins, L.M, Wilson, O, Snow, M.E, Wang, Y.-S. | | Deposit date: | 1997-06-12 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the Src homology domain-2 of the growth factor receptor-bound protein-2.
J.Biomol.NMR, 11, 1998
|
|
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
4TQF
 
 | | Crystal Structure of the C-terminal domain of IFRS bound with 2-(5-bromothienyl)-L-Ala and ATP | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-bromothiophen-2-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, A, O'Donoghue, P, Soll, D. | | Deposit date: | 2014-06-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7143 Å) | | Cite: | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5E
 
 | | Structure of BACE Bound to SCH736062 | | Descriptor: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5D
 
 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5F
 
 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
8H5B
 
 | | The cryo-EM structure of nuclear transport receptor Kap114p complex with yeast TATA-box binding protein | | Descriptor: | Importin subunit beta-5, TATA-box-binding protein | | Authors: | Hsia, K.C, Liao, C.C, Wang, C.H, Wu, Y.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural convergence endows nuclear transport receptor Kap114p with a transcriptional repressor function toward TATA-binding protein.
Nat Commun, 14, 2023
|
|
5UD5
 
 | |
5V6X
 
 | |
