6AC6
 
 | | Ab initio crystal structure of Selenomethionine labelled Mycobacterium smegmatis Mfd | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACA
 
 | | Crystal structure of Mycobacterium tuberculosis Mfd at 3.6 A resolution | | Descriptor: | Mycobacterium tuberculosis Mfd | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6AC8
 
 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACX
 
 | | Crystal structure of Mycobacterium smegmatis Mfd in complex with ADP + Pi at 3.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mycobacterium smegmatis Mfd, PHOSPHATE ION, ... | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
5G60
 
 | | S.pneumoniae ABC-transporter substrate binding protein FusA in complex with nystose | | Descriptor: | ABC TRANSPORTER, SUBSTRATE-BINDING PROTEIN, CALCIUM ION, ... | | Authors: | Culurgioni, S, Harris, G, Singh, A.K, King, S.J, Walsh, M.A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Regulation and Specificity of Fructooligosaccharide Import in Streptococcus pneumoniae.
Structure, 25, 2017
|
|
5G5Y
 
 | | S.pneumoniae ABC-transporter substrate binding protein FusA apo structure | | Descriptor: | ABC TRANSPORTER, SUBSTRATE-BINDING PROTEIN, CALCIUM ION, ... | | Authors: | Culurgioni, S, Harris, G, Singh, A.K, King, S.J, Walsh, M.A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for Regulation and Specificity of Fructooligosaccharide Import in Streptococcus pneumoniae.
Structure, 25, 2017
|
|
1OL0
 
 | | Crystal structure of a camelised human VH | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN G, SULFATE ION | | Authors: | Dottorini, T, Vaughan, C.K, Walsh, M.A, Losurdo, P, Sollazzo, M. | | Deposit date: | 2003-08-02 | | Release date: | 2004-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Human Vh: Requirements for Maintaining a Monomeric Fragment
Biochemistry, 43, 2004
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
6RAB
 
 | |
6RD1
 
 | | Ruminococcus gnavus sialic acid aldolase catalytic lysine mutant in complex with sialic acid | | Descriptor: | 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, Putative N-acetylneuraminate lyase | | Authors: | Owen, C.D, Bell, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-04-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.
Nat Microbiol, 4, 2019
|
|
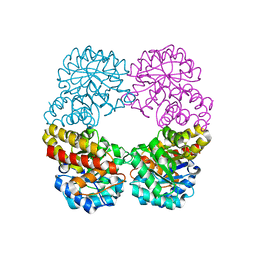
6RB7
 
 | | Ruminococcus gnavus sialic acid aldolase catalytic lysine mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Bell, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-04-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.
Nat Microbiol, 4, 2019
|
|
7OFZ
 
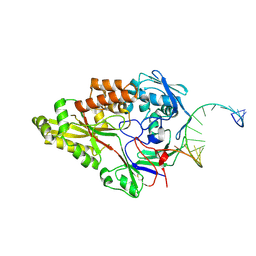 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OG0
 
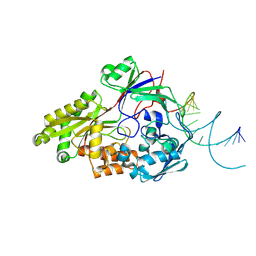 | | Nontypeable Haemophillus influenzae SapA in open and closed conformations, in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
5O7O
 
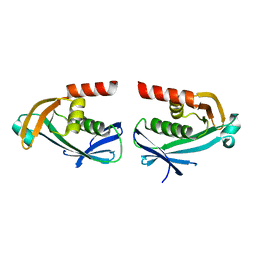 | | The crystal structure of DfoC, the desferrioxamine biosynthetic pathway acetyltransferase/Non-Ribosomal Peptide Synthetase (NRPS)-Independent Siderophore (NIS) from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | Desferrioxamine siderophore biosynthesis protein dfoC | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O3Z
 
 | | Crystal structure of Sorbitol-6-Phosphate 2-dehydrogenase SrlD from Erwinia amylovora | | Descriptor: | CHLORIDE ION, Sorbitol-6-phosphate dehydrogenase | | Authors: | Salomone-Stagni, M, Bartho, J.D, Bellini, D, Walsh, M.A, Benini, S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and functional analysis of Erwinia amylovora SrlD. The first crystal structure of a sorbitol-6-phosphate 2-dehydrogenase.
J.Struct.Biol., 203, 2018
|
|
5O8R
 
 | | The crystal structure of DfoA bound to FAD and NADP; the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O8P
 
 | | The crystal structure of DfoA bound to FAD, the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J.Struct.Biol., 202, 2018
|
|
4MDZ
 
 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FE (III) ION, Metal dependent phosphohydrolase, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-23 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
7OFW
 
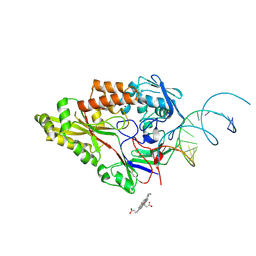 | | Nontypeable Haemophillus influenzae SapA in complex with heme | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
4ME4
 
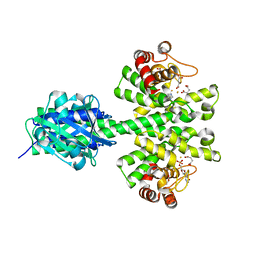 | |
9FGO
 
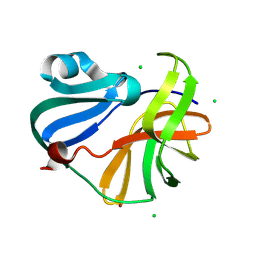 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
5M1T
 
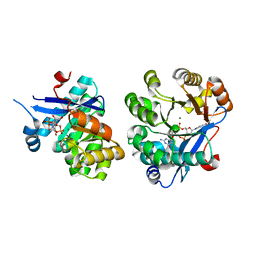 | | PaMucR Phosphodiesterase, c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, MucR Phosphodiesterase | | Authors: | Hutchin, A, Tews, I, Walsh, M.A. | | Deposit date: | 2016-10-10 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
4MCW
 
 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, IMIDAZOLE, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-21 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
5O5C
 
 | | The crystal structure of DfoJ, the desferrioxamine biosynthetic pathway lysine decarboxylase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
