3QNY
 
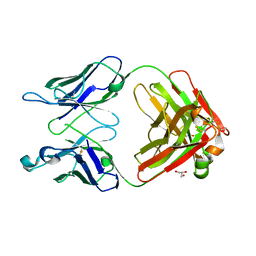 | | Monoclinic form of human IgA1 Fab fragment, sharing same Fv as IgG | | Descriptor: | FAB FRAGMENT OF IMMUNOGLOBULIN A1 HEAVY CHAIN, FAB FRAGMENT OF IMMUNOGLOBULIN A1 LIGHT CHAIN, GLYCEROL | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QNX
 
 | | Orthorhombic form of human IgA1 Fab fragment, sharing same Fv as IgG | | Descriptor: | Fab fragment of IMMUNOGLOBULIN A1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN A1 LIGHT CHAIN, GLYCEROL | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QNZ
 
 | | Orthorhombic form of IgG1 Fab fragment (in complex with antigenic tubulin peptide) sharing same Fv as IgA | | Descriptor: | Fab fragment of IMMUNOGLOBULIN G1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN G1 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QO1
 
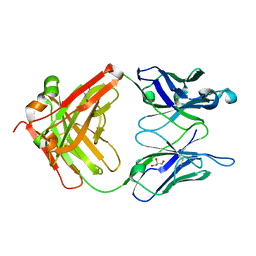 | | Monoclinic form of IgG1 Fab fragment (apo form) sharing same Fv as IgA | | Descriptor: | Fab fragment of IMMUNOGLOBULIN G1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN G1 LIGHT CHAIN, GLYCEROL | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | Descriptor: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | Authors: | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.83 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
6NQW
 
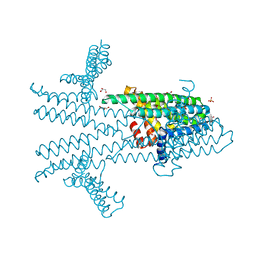 | | Flagellar protein FcpA from Leptospira biflexa - hexagonal form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Flagellar coiling protein A, GLYCEROL, ... | | Authors: | San Martin, F, Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
6NQY
 
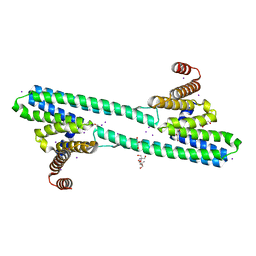 | | Flagellar protein FcpA from Leptospira biflexa / ab-centered monoclinic form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Flagellar coiling protein A, ... | | Authors: | Mechaly, A, Larrieux, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete.
Elife, 9, 2020
|
|
6NQX
 
 | | Flagellar protein FcpA from Leptospira biflexa / primitive monoclinic form | | Descriptor: | Flagellar coiling protein A, GLYCEROL | | Authors: | Mechaly, A, Larrieux, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
6XQK
 
 | | Crystal structure of the D/D domain of PKA from S. cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, cAMP-dependent protein kinase regulatory subunit | | Authors: | Larrieux, N, Gonzalez Bardeci, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of yeast regulatory subunit reveals key evolutionary insights into Protein Kinase A oligomerization.
J.Struct.Biol., 213, 2021
|
|
5CEE
 
 | | Malic enzyme from Candidatus Phytoplasma AYWB in complex with NAD and Mg2+ | | Descriptor: | MAGNESIUM ION, NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Alvarez, C.E, Trajtenberg, F, Larrieux, N, Saigo, M, Mussi, M.A, Andreo, C.S, Drincovich, M.F, Buschiazzo, A. | | Deposit date: | 2015-07-06 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3M8O
 
 | | Human IgA1 Fab fragment | | Descriptor: | CHLORIDE ION, GLYCEROL, IMMUNOGLOBULIN A1 HEAVY CHAIN, ... | | Authors: | Buschiazzo, A, Trajtenberg, F, Correa, A, Oppezzo, P, Pritsch, O, Dighiero, G. | | Deposit date: | 2010-03-18 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4EMX
 
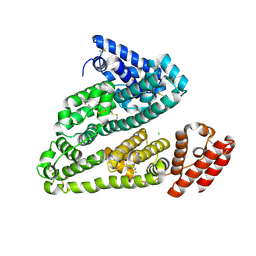 | | Crystal structure analysis of Human Serum Albumin in complex with chloride anions at cryogenic temperature | | Descriptor: | CHLORIDE ION, Serum albumin | | Authors: | Botti, H, Bonilla, L, Trajtenberg, F, Radi, R, Buschiazzo, A. | | Deposit date: | 2012-04-12 | | Release date: | 2012-04-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights on B factors in crystal structure analysis and crystallographic model refinement
to be published
|
|
6O6P
 
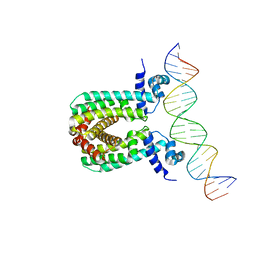 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with DNA | | Descriptor: | DNA-forward, DNA-reverse, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.851 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6OC8
 
 | |
6O6O
 
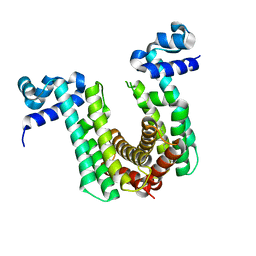 | | Structure of the regulator FasR from Mycobacterium tuberculosis | | Descriptor: | MYRISTIC ACID, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6O6N
 
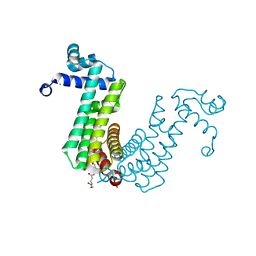 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with C20-CoA | | Descriptor: | Arachinoyl-CoA, CHLORIDE ION, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
5OU5
 
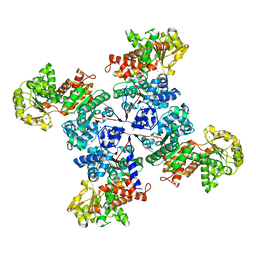 | |
3EHJ
 
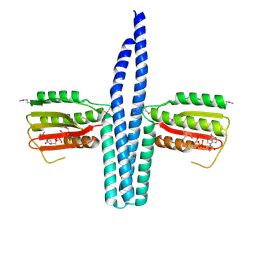 | |
3EHH
 
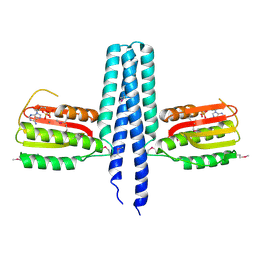 | |
3EHF
 
 | |
