8BFJ
 
 | | CNOT11-GGNBP2 complex | | Descriptor: | 1,2-ETHANEDIOL, CCR4-NOT transcription complex subunit 11, Gametogenetin-binding protein 2 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
6Y3P
 
 | |
6Y3Z
 
 | |
2HYI
 
 | | Structure of the human exon junction complex with a trapped DEAD-box helicase bound to RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*U)-3', MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andersen, C.B.F, Le Hir, H, Andersen, G.R. | | Deposit date: | 2006-08-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
3P26
 
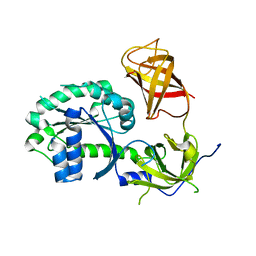 | | Crystal structure of S. cerevisiae Hbs1 protein (apo-form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3P27
 
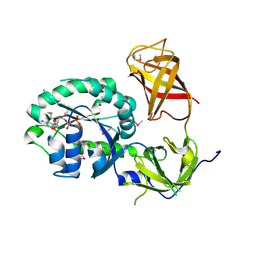 | | Crystal structure of S. cerevisiae Hbs1 protein (GDP-bound form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5LSL
 
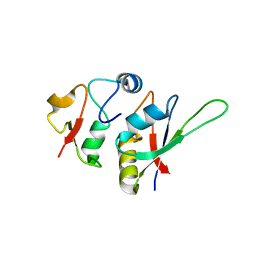 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-09-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
5LSB
 
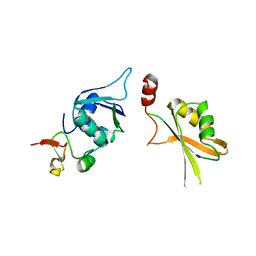 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
4B8A
 
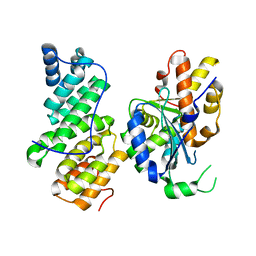 | | Structure of yeast NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8B
 
 | | N-Terminal domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GOLD ION | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B89
 
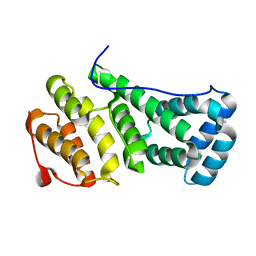 | | MIF4G domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8C
 
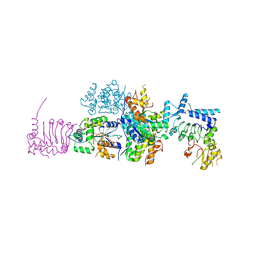 | | nuclease module of the yeast Ccr4-Not complex | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GLUCOSE-REPRESSIBLE ALCOHOL DEHYDROGENASE TRANSCRIPTIONAL EFFECTOR, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4A8J
 
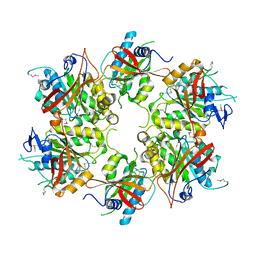 | | Crystal Structure of the Elongator subcomplex Elp456 | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Elongator Subcomplex Elp456 is a Hexameric Reca-Like ATPase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4BY6
 
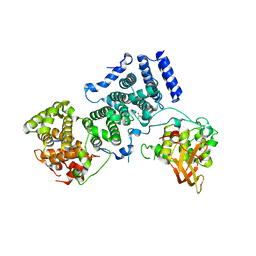 | | Yeast Not1-Not2-Not5 complex | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure and RNA-Binding Properties of the not1-not2-not5 Module of the Yeast Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
5L7L
 
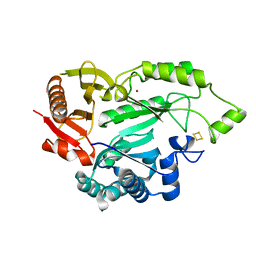 | |
5M2N
 
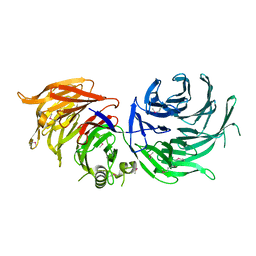 | |
5L7J
 
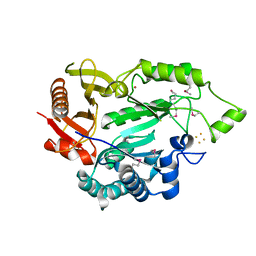 | |
2JE6
 
 | | Structure of a 9-subunit archaeal exosome | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2007-01-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Channelling by the Archaeal Exosome.
Embo Rep., 8, 2007
|
|
2JEA
 
 | |
2JEB
 
 | |
4WZJ
 
 | | Spliceosomal U4 snRNP core domain | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Leung, A.K.W, Nagai, K, Li, J. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
