5MFM
 
 | | Designed armadillo repeat protein peptide fusion YIIIM6AII_GS11_(KR)5 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Importin subunit alpha, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFF
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, 1,2-ETHANEDIOL, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFN
 
 | | Designed armadillo repeat protein YIIIM5AII | | Descriptor: | CALCIUM ION, D-MALATE, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MAD
 
 | | GFP-binding DARPin 3G61 | | Descriptor: | 3G61, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFB
 
 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFH
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
8BGM
 
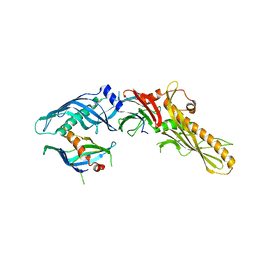 | |
4DXZ
 
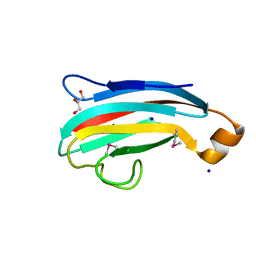 | | crystal structure of a PliG-Ec mutant, a periplasmic lysozyme inhibitor of g-type lysozyme from Escherichia coli | | Descriptor: | GLYCEROL, Inhibitor of g-type lysozyme, SODIUM ION | | Authors: | Leysen, S, Vanheuverzwijn, S, Van Asten, K, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | crystal structure of a PliG-Ec mutant, a periplasmic lysozyme inhibitor of g-type lysozyme from Escherichia coli
To be Published
|
|
7QFQ
 
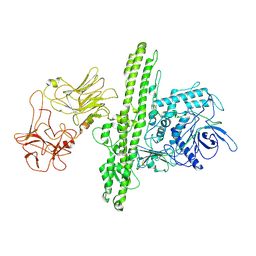 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7QFP
 
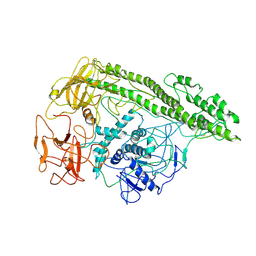 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
5MA3
 
 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA5
 
 | | GFP-binding DARPin fusion gc_K11 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MA9
 
 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
8CEY
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-(3,3-dimethylcyclobutyl)imidazo[5,1-b][1,3]thiazole-3-carboxamide | | Authors: | Kosenina, S, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233
To Be Published
|
|
8CEX
 
 | |
5MFG
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)4 | | Descriptor: | (RR)4, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFI
 
 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFK
 
 | | Designed armadillo repeat protein YIII(Dq.V1)4CPAF in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V1)4CPAF | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
4G9S
 
 | | Crystal structure of Escherichia coli PliG in complex with Atlantic salmon g-type lysozyme | | Descriptor: | CHLORIDE ION, CITRATE ANION, Goose-type lysozyme, ... | | Authors: | Leysen, S, Vanderkelen, L, Weeks, S.D, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-07-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis of bacterial defense against g-type lysozyme-based innate immunity.
Cell.Mol.Life Sci., 70, 2013
|
|
6QIU
 
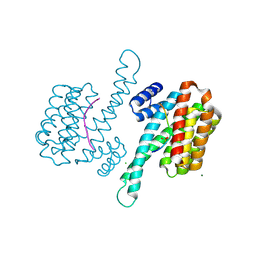 | | Crystal structure of 14-3-3 sigma in complex with Ataxin-1 Ser776 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Ataxin-1 phosphopeptide, CHLORIDE ION, ... | | Authors: | Leysen, S, Milroy, L.G, Davis, J.M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2019-01-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1
To Be Published
|
|
3OE3
 
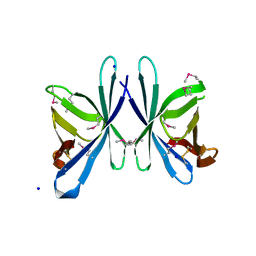 | | Crystal structure of PliC-St, periplasmic lysozyme inhibitor of C-type lysozyme from Salmonella typhimurium | | Descriptor: | Putative periplasmic protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-12 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
3P53
 
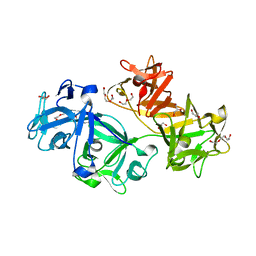 | | Structure of fascin | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DODECAETHYLENE GLYCOL, Fascin, ... | | Authors: | Jansen, S, Dominguez, R. | | Deposit date: | 2010-10-07 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of actin filament bundling by fascin.
J.Biol.Chem., 286, 2011
|
|
3PTM
 
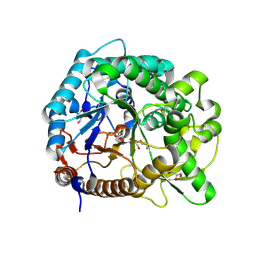 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with 2-fluoroglucopyranoside | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
3PTK
 
 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase Os4BGlu12, ZINC ION | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
5AEI
 
 | | Designed Armadillo repeat protein YIIIM5AII in complex with peptide (KR)5 | | Descriptor: | ACETATE ION, CALCIUM ION, DESIGNED ARMADILLO REPEAT PROTEIN YIIIM5AII, ... | | Authors: | Hansen, S, Tremmel, D, Madhurantakam, C, Reichen, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2015-08-31 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Energetic Contributions of a Designed Modular Peptide-Binding Protein with Picomolar Affinity.
J.Am.Chem.Soc., 138, 2016
|
|
