3CQ2
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Ebihara, A, Chen, L, Liu, Z.J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein from Thermus Thermophilus HB8
To be Published
|
|
3CPQ
 
 | | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044) | | Descriptor: | 50S ribosomal protein L30e | | Authors: | Jeyakanthan, J, Sarani, R, Mridula, P, Sekar, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L30e a ribosomal protein from Methanocaldococcus jannaschii DSM2661 (MJ1044)
To be Published
|
|
5XB3
 
 | |
5X8D
 
 | |
5X8K
 
 | |
5X8B
 
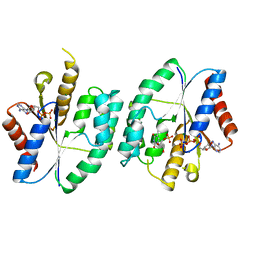 | | Crystal structure of ATP-TMP and ADP bound thymidylate kinase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5X8V
 
 | |
5X98
 
 | |
5XBH
 
 | |
5X86
 
 | | Crystal structure of TMP bound thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5XAL
 
 | |
5XT8
 
 | | Magnesium bound apo structure of thymidylate kinase (form I) from Thermus thermophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-06-17 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5X7J
 
 | | Crystal structure of thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2019
|
|
5XB5
 
 | |
5XB2
 
 | | ADP-Mg-F-dTMP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate
FEBS J., 284, 2017
|
|
5XAI
 
 | |
5XAK
 
 | |
7WRK
 
 | |
7WWO
 
 | |
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
5H4G
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 1.77 A resolution | | Descriptor: | Ribonuclease VapC4, ZINC ION | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4H
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 2.2 A resolution | | Descriptor: | CADMIUM ION, Ribonuclease VapC4 | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4F
 
 | | Structure of inorganic pyrophosphatase crystallised as a contaminant | | Descriptor: | ZINC ION, inorganic pyrophosphatase | | Authors: | Chaudhary, S, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
474D
 
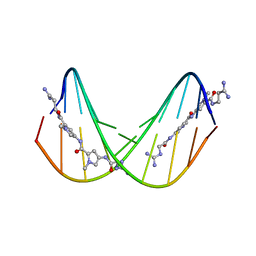 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
1UE7
 
 | | Crystal structure of the single-stranded dna-binding protein from mycobacterium tuberculosis | | Descriptor: | Single-strand binding protein | | Authors: | Saikrishnan, K, Jeyakanthan, J, Venkatesh, J, Acharya, N, Sekar, K, Varshney, U, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-09 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications
J.MOL.BIOL., 331, 2003
|
|
