9IXO
 
 | | Crystal structure of OXA-14 | | Descriptor: | Beta-lactamase | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXP
 
 | | Crystal structure of OXA-10 variant A124T | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXR
 
 | | Crystal structure of OXA-10 variant A124T in the complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXN
 
 | | Crystal structure of OXA-10 | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
9IXQ
 
 | | Crystal structure of OXA-17 | | Descriptor: | Beta-lactamase | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
4EDA
 
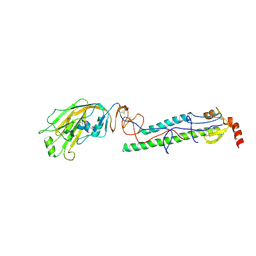 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
6FWZ
 
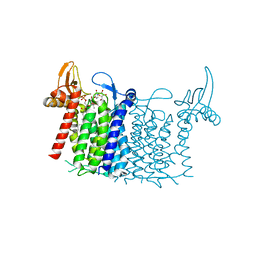 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-07 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
6FM9
 
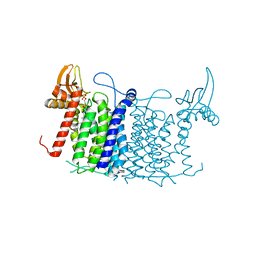 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
3ZHK
 
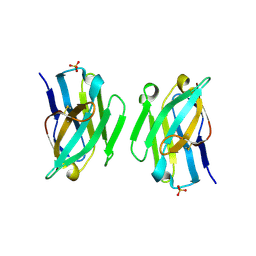 | |
3ZHD
 
 | |
3ZHL
 
 | |
3LDI
 
 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | GLYCEROL, MERCURY (II) ION, Pancreatic trypsin inhibitor, ... | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3LDJ
 
 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, Pancreatic trypsin inhibitor | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding.
Biochem.Biophys.Res.Commun., 2010
|
|
3LDM
 
 | |
5LEV
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) | | Descriptor: | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, UNKNOWN LIGAND | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
7MJR
 
 | | Vip4Da2 toxin structure | | Descriptor: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | Authors: | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
6ON6
 
 | | Crystal Structure of the RIG-5 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NeuRonal IgCAM-5 | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PLL
 
 | | Crystal structure of the ZIG-8 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Zwei Ig domain protein zig-8 | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6POL
 
 | | Crystal structure of the human NELL1 EGF1-3-Robo3 FN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
6POK
 
 | | Crystal structure of the Robo3 FN2-3 domains | | Descriptor: | GLYCEROL, Roundabout homolog 3 | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
6POG
 
 | | Crystal structure of the NELL2 EGF1-6-Robo3 FN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-03 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
6PWN
 
 | | MscS Nanodisc with N-terminal His-Tag | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, HEXADECANE, Small-conductance mechanosensitive channel | | Authors: | Reddy, B.G, Perozo, E. | | Deposit date: | 2019-07-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS.
Elife, 8, 2019
|
|
6PWO
 
 | | MscS DDM | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Reddy, B.G, Perozo, E. | | Deposit date: | 2019-07-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS.
Elife, 8, 2019
|
|
