2LAB
 
 | |
2PA2
 
 | | Crystal structure of human Ribosomal protein L10 core domain | | Descriptor: | 60S ribosomal protein L10, POTASSIUM ION | | Authors: | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
1IS1
 
 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
1DRN
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GGAGA)R(UGAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*GP*AP*GP*A)-R(P*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-11-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
1DHH
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GG)R(AGAU)D(GAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*G)-R(P*AP*GP*AP*U)-D(P*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-10-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
6KQ1
 
 | | Crystal structure of cytochrome c551 from Pseudomonas sp. strain MT-1. | | Descriptor: | Cytochrome C biogenesis protein CcsA, HEME C, ZINC ION | | Authors: | Fujii, S, Oki, H, Kawahara, K, Ohkubo, T, Masanari-Fujii, M, Wakai, S, Sambongi, Y. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into high stability of cytochrome c551 from a deep-sea piezo-tolerant bacterium, Pseudomonas sp. strain MT-1
To Be Published
|
|
3WJ4
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WJ5
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with triphenyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, triphenylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
1VA2
 
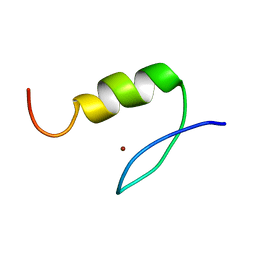 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
1VA3
 
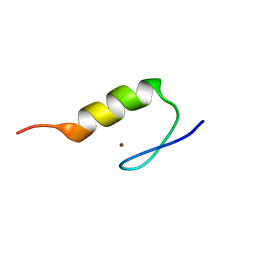 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
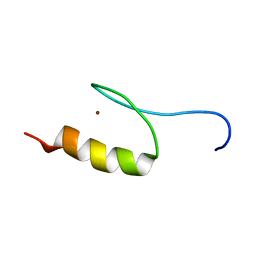 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1IWC
 
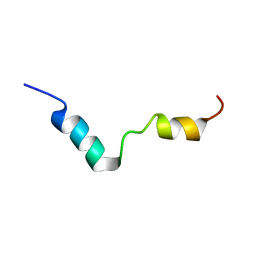 | | TFE-induded structure of the N-terminal domain of pig gastric H/K-ATPase | | Descriptor: | gastric H/K-ATPase | | Authors: | Fujitani, N, Kanagawa, M, Aizawa, T, Ohkubo, T, Kaya, S, Demura, M, Kawano, K, Taniguchi, K, Nitta, K. | | Deposit date: | 2002-05-02 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure determination and conformational change induced by tyrosine phosphorylation of the N-terminal domain of the alpha-chain of pig gastric H+/K+-ATPase
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1ISE
 
 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
1IWF
 
 | | Solution structure of the N-terminal domain of pig gastric H/K-ATPase | | Descriptor: | gastric H/K-ATPase | | Authors: | Fujitani, N, Kanagawa, M, Aizawa, T, Ohkubo, T, Kaya, S, Demura, M, Kawano, K, Taniguchi, K, Nitta, K. | | Deposit date: | 2002-05-06 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure determination and conformational change induced by tyrosine phosphorylation of the N-terminal domain of the alpha-chain of pig gastric H+/K+-ATPase
Biochem.Biophys.Res.Commun., 300, 2003
|
|
6A3L
 
 | | Crystal structure of cytochrome c' from Shewanella violacea DSS12 | | Descriptor: | Cytochrome c, HEME C | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6A3K
 
 | | Crystal structure of cytochrome c' from Shewanella benthica DB6705 | | Descriptor: | Cytochrome c, HEME C, PENTAETHYLENE GLYCOL | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
1WKI
 
 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
1Y1C
 
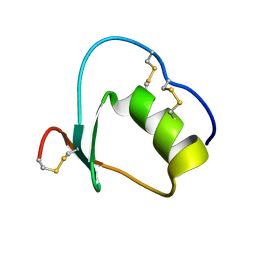 | | Solution structure of Anemonia elastase inhibitor analogue | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
1Y1B
 
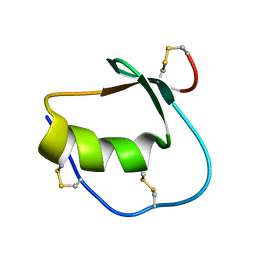 | | Solution structure of Anemonia elastase inhibitor | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1996-09-25 | | Release date: | 1997-10-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
1GCF
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, 12 STRUCTURES | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1997-04-10 | | Release date: | 1997-10-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
1WZB
 
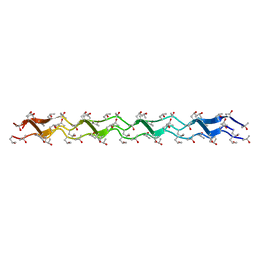 | | Crystal structure of the collagen triple helix model [{HYP(R)-HYP(R)-GLY}10]3 | | Descriptor: | Collagen triple helix | | Authors: | Kawahara, K, Nakamura, S, Nishi, Y, Uchiyama, S, Nishiuchi, Y, Nakazawa, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of hydration on the stability of the collagen-like triple-helical structure of [4(R)-hydroxyprolyl-4(R)-hydroxyprolylglycine]10
Biochemistry, 44, 2005
|
|
