1WNG
 
 | |
1WM6
 
 | |
3VKM
 
 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
2ZHM
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 1) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHL
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 2) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHK
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 1) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHN
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 2) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, GALECTIN-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2D6P
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with T-antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6O
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6L
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD (crystal form 2) | | Descriptor: | lectin, galactose binding, soluble 9 | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6M
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6K
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD (crystal form 1) | | Descriptor: | lectin, galactose binding, soluble 9 | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6N
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2EAL
 
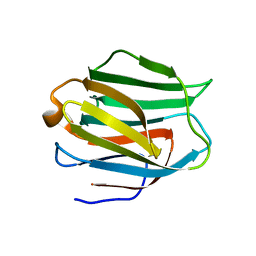 | | Crystal structure of human galectin-9 N-terminal CRD in complex with Forssman pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, Galectin-9 | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
2EAK
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, Galectin-9, ... | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
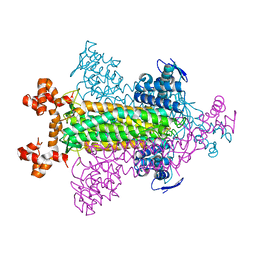
1VDK
 
 | |
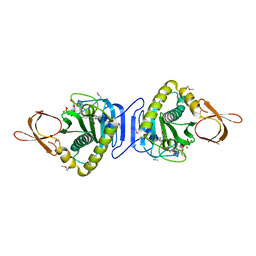
2FYK
 
 | |
1ARU
 
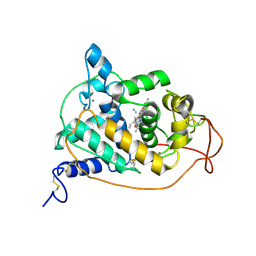 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARV
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARW
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARX
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARY
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ISR
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with Glutamate and Gadolinium Ion | | Descriptor: | GADOLINIUM ATOM, GLUTAMIC ACID, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ISS
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with an antagonist | | Descriptor: | (S)-(ALPHA)-METHYL-4-CARBOXYPHENYLGLYCINE, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1AF3
 
 | | RAT BCL-XL AN APOPTOSIS INHIBITORY PROTEIN | | Descriptor: | APOPTOSIS REGULATOR BCL-X | | Authors: | Aritomi, M, Kunishima, N, Inohara, N, Ishibashi, Y, Ohta, S, Morikawa, K. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat Bcl-xL. Implications for the function of the Bcl-2 protein family.
J.Biol.Chem., 272, 1997
|
|
