6GV9
 
 | | Structure of the type IV pilus from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Bardiaux, B, Amorim, G.C, Luna-Rico, A, Zheng, W, Guilvout, I, Jollivet, C, Nilges, M, Egelman, E, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
1AXH
 
 | | ATRACOTOXIN-HVI FROM HADRONYCHE VERSUTA (AUSTRALIAN FUNNEL-WEB SPIDER, NMR, 20 STRUCTURES | | Descriptor: | ATRACOTOXIN-HVI | | Authors: | Fletcher, J.I, O'Donoghue, S.I, Nilges, M, King, G.F. | | Deposit date: | 1996-11-04 | | Release date: | 1997-11-12 | | Last modified: | 2021-02-03 | | Method: | SOLUTION NMR | | Cite: | The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider.
Nat.Struct.Biol., 4, 1997
|
|
1AOY
 
 | |
1M7L
 
 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
1BPV
 
 | |
1MEK
 
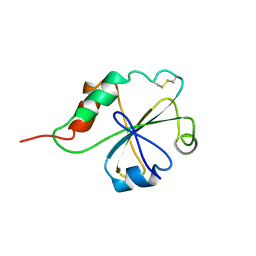 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 40 STRUCTURES | | Descriptor: | PROTEIN DISULFIDE ISOMERASE | | Authors: | Kemmink, J, Darby, N.J, Dijkstra, K, Nilges, M, Creighton, T.E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-04-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy.
Biochemistry, 35, 1996
|
|
2GVB
 
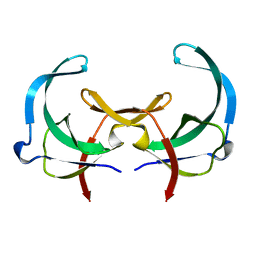 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
2GVA
 
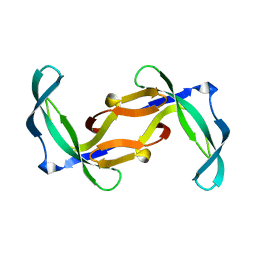 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
1ZBJ
 
 | |
1VIH
 
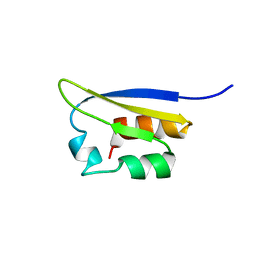 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VIG
 
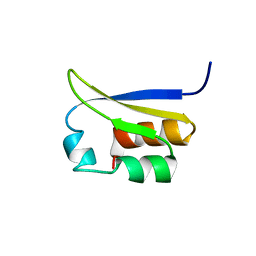 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1QXN
 
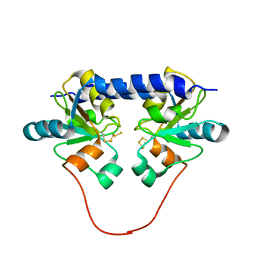 | | Solution Structure of the 30 kDa Polysulfide-sulfur Transferase Homodimer from Wolinella Succinogenes | | Descriptor: | PENTASULFIDE-SULFUR, sulfide dehydrogenase | | Authors: | Lin, Y.J, Dancea, F, Loehr, F, Klimmek, O, Pfeiffer-Marek, S, Nilges, M, Wienk, H, Kroeger, A, Rueterjans, H. | | Deposit date: | 2003-09-08 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 30 kDa Polysulfide-Sulfur Transferase Homodimer from Wolinella succinogenes
Biochemistry, 43, 2004
|
|
2J0E
 
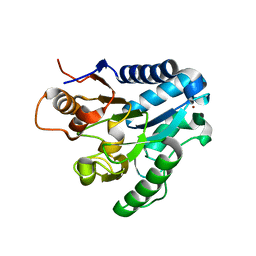 | | Three dimensional structure and catalytic mechanism of 6- phosphogluconolactonase from Trypanosoma brucei | | Descriptor: | 6-PHOSPHOGLUCONOLACTONASE, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Delarue, M, Duclert-Savatier, N, Miclet, E, Haouz, A, Giganti, D, Ouazzani, J, Lopez, P, Nilges, M, Stoven, V. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Dimensional Structure and Implications for the Catalytic Mechanism of 6-Phosphogluconolactonase from Trypanosoma Brucei.
J.Mol.Biol., 366, 2007
|
|
2FMR
 
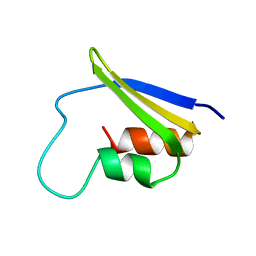 | | KH1 FROM THE FRAGILE X PROTEIN FMR1, NMR, 18 STRUCTURES | | Descriptor: | FMR1 PROTEIN | | Authors: | Musco, G, Kharrat, A, Stier, G, Fraternali, F, Gibson, T.J, Nilges, M, Pastore, A. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome.
Nat.Struct.Biol., 4, 1997
|
|
1PFS
 
 | | SOLUTION NMR STRUCTURE OF THE SINGLE-STRANDED DNA BINDING PROTEIN OF THE FILAMENTOUS PSEUDOMONAS PHAGE PF3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PF3 SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Folmer, R.H.A, Nilges, M, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1996-08-03 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the single-stranded DNA binding protein of the filamentous Pseudomonas phage Pf3: similarity to other proteins binding to single-stranded nucleic acids.
EMBO J., 14, 1995
|
|
2KUB
 
 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
1UL9
 
 | | CGL2 ligandfree | | Descriptor: | galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULC
 
 | | CGL2 in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULF
 
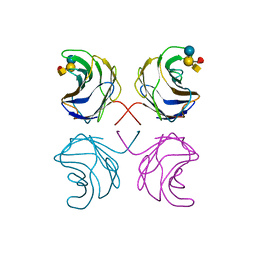 | | CGL2 in complex with Blood Group A tetrasaccharide | | Descriptor: | alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULE
 
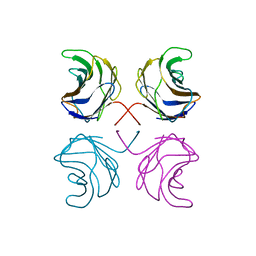 | | CGL2 in complex with linear B2 trisaccharide | | Descriptor: | alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULD
 
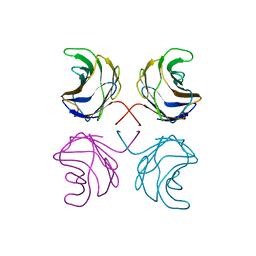 | | CGL2 in complex with blood group H type II | | Descriptor: | alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULG
 
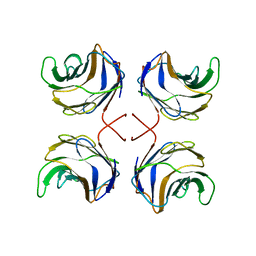 | | CGL2 in complex with Thomsen-Friedenreich antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
3E7F
 
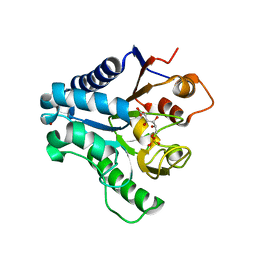 | | Crystal structure of 6-phosphogluconolactonase from Trypanosoma brucei complexed with 6-phosphogluconic acid | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconolactonase, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-18 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
3EB9
 
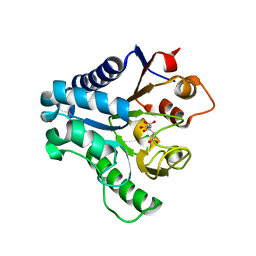 | | Crystal structure of 6-phosphogluconolactonase from trypanosoma brucei complexed with citrate | | Descriptor: | 6-phosphogluconolactonase, CITRATE ANION, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-27 | | Release date: | 2009-05-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
1ATE
 
 | |
